Introduction
Cervical cancer is the second most common type of
cancer in females worldwide and is a leading cause of
cancer-related mortality in females in developing countries.
Despite the public health importance of cervical cancer in China,
cervical cancer is most common among females, followed by breast
cancer, with an estimated 1,520,000 new cases and 780,000
fatalities (1). Multiple
epidemiological studies have indicated that the human
papillomavirus infection is frequently detected in invasive
cervical cancer (2). However, this
alone is not sufficient for malignant transformation and other
genetic events are required for the development of cancer. The
genetic basis underlying cervical tumorigenesis and progression are
largely unknown. Recently, much progress has been made in
identifying the genes that are involved in the development of
cervical cancer, which is useful for understanding the pathogenesis
of cervical cancer and defining its molecular signature (3–6).
Phosphatase and tensin homologue (PTEN) is a tumor
suppressor gene that is located on human chromosome band 10q23.31,
contains 9 exons and encodes a 403 amino acid sequence for a
dual-specificity phosphatase with homology to the cytoskeleton
proteins, chicken tensin and bovine auxilin, which interact with
adhesion molecules (7–10). The mutations of the PTEN gene have
been found in a variety of human cancer cell lines and primary
tumors of the brain, stomach, breast and kidney in different
frequencies (11–15), indicating that the gene may play a
role in the tumorigenesis of certain cancers. PTEN promoter
methylation and reduced PTEN expression were also found in gastric,
non-small cell lung and cervical cancers (16–18).
However, little is known regarding the association between
mutation, promoter methylation and the PTEN gene expression in
cervical cancer in the Chinese population. Therefore, the aim of
the present study was to investigate the function of PTEN in
cervical cancer and to correlate it further with
clinicopathological characteristics to determine its clinical
importance.
Materials and methods
Tissue samples
Between January 2011 and April 2012, 102 paraffin
blocks of consecutive patients with cervical cancer who underwent a
cervical biopsy at the Department of Surgery of Suzhou University,
Affiliated Changzhou Tumor Hospital (Jiangsu, China) were enrolled
in the study. None of the patients received preoperative
chemotherapy and/or radiation therapy. The patients whose surgical
tissue was studied provided their written informed consent.
Histological studies were also performed and all the specimens were
characterized as cervical cancer. The Bioethics Committee of the
Institution approved the study. The histological types and grades
of tumor were classified according to World Health Organization
criteria. The clinical staging of each tumor was established
according to the International Federation of Gynecology and
Obstetrics (FIGO) criteria.
DNA extraction
Formalin-fixed paraffin samples were cut into 10-μm
sections. The sections were pulverized under liquid nitrogen using
a Mikro-Dismembrator (B. Braun Biotech International, Melsungen,
Germany). For each sample, 0.1 g of pulverized tissue powder was
resuspended in 1 ml of 0.25% xylene and was maintained for 15 min
at 55°C. The suspension was centrifuged at 14,000 × g for 5 min and
the pellet was suspended in 0.1 ml of xylene and processed as
aforementioned for a second time. The resulted sediment was mixed
with 100% ethanol and processed with xylene lysis buffer [(Tris,
sodium dodecyl sulfate and ethylenediaminetetraacetic acid (EDTA)].
A lysis buffer containing 300 μg/ml of proteinase K was added to
the pellet, mixed and incubated at 55°C overnight. The DNA was
extracted by the phenol-chloroform procedure and subsequently
dissolved in TE buffer (Tris-HCl and EDTA) and stored at −20°C.
Polymerase chain reaction-single-strand
conformational polymorphism (PCR-SSCP)
Tumor DNA was subjected to PCR amplification. All 9
exons of the PTEN gene were amplified using primers designed and
synthesized by Invitrogen Corp. (Shanghai, China) (Table I). PCR amplification was performed
within a reaction mixture containing 2.5 μl of 10× PCR buffer
(Mg2+-free; Takara Bio, Inc., Shiga, Japan), 0.8 μl
Mg2+ (50 mM), 0.5 μl dNTP (10 mM, Takara), 0.5 μl primer
F (10 μm), 0.5 μl primer R (10 μm), 0.2 μl Taq polymerase (5 U/μl,
Takara Bio, Inc.,), 1.0 μl genomic DNA and 19.0 μl
ddH2O. A programmable thermocycler (Eppendorf PCR system
9700, Hamburg, Germany) was used to perform the amplifications. The
PCR condition for each exon was as follows: Pre-denaturation for 5
min at 95°C, 35 cycles of 30 sec denaturation at 95°C, 40 sec
annealing at the temperature shown in Table I, 60 sec extension at 72°C and 10
min final extension at 72°C. The amplified PCR products were
denatured at 95°C for 5 min and run on an 8% denaturation
polyacrylamide gel (1× Tris/Borate/EDTA buffer, circulatory water)
at a voltage of 100 V and at 10°C for 16 h. Silver staining was
performed as previously described (19). According to the PCR-SSCP results of
the genome DNA, the difference in the single-strand strip number
and electrophoresis transference location, also known as the
mobility shift, was considered as PCR-SSCP positive. The genome DNA
from the positive-PCR-SSCP samples was amplified for bidirectional
DNA sequencing. The amplified PCR products were sequenced with an
ABI Prism 310 Dye Terminator Cycle Sequencing ready reaction kit
(Applied Biosystems, Foster City, CA, USA).
 | Table IOligonucleotide primers for
phosphatase and tensin homologue analysis. |
Table I
Oligonucleotide primers for
phosphatase and tensin homologue analysis.
| Exon | Forward (5′-3′) | Reverse (5′-3′) | Annealing temp.
(°C) | Product size
(bp) |
|---|
| 1 |
TCCTCCTTTTTCTTCAGCCAC |
GAAAGGTAAAGAGGAGCAGCC | 56 | 147 |
| 2 |
TGCATATTTCAGATATTTCTTTCCTT |
TTTGAAATAGAAAATCAAAGCATTC | 57 | 155 |
| 3 |
TGTTAATGGTGGCTTTTTG |
GCAAGCATACAAATAAGAAAAC | 56 | 114 |
| 4 |
TTCCTAAGTGCAAAAGATAAC |
TACAGTCTATCGGGTTTAAGT | 56 | 147 |
| 5 |
TTTTTTTTTCTTATTCTGAGGTTATC |
GAAGAGGAAAGGAAAAACATC | 51 | 312 |
| 6 |
AGTGAAATAACTATAATGGAACA |
GAAGGATGAGAATTTCAAGC | 54 | 231 |
| 7 |
ATCGTTTTTGACAGTTTG |
TCCCAATGAAAGTAAAGTAGA | 55 | 262 |
| 8 |
AGGTGACAGATTTTCTTTTTTA |
TAGCTGTACTCCTAGAATTA | 52 | 394 |
| 9 |
GTTCATCTGCAAAATGGA |
TGGTAATCTGACACAATGTCCTA | 50 | 397 |
Promoter methylation analysis
Promoter methylation of the PTEN gene was determined
by methylation-specific-PCR (MS-PCR). The genomic DNA from
carcinoma specimens was subjected to bisulfite modifications using
the EZ DNA Methylation-Gold™ kit (Zymo Research, Irvine, CA, USA)
according to the manufacturer’s instructions. The modified DNA was
used as a template for MS-PCR as described above. The primers for
the unmethylated PTEN were forward, 5′-TTTTTTGGGTTTTTGAAATTTAATG-3′
and reverse, 5′-AACCTACTATTATATCACCAACATA-3′ yielding a 165-bp
product. The primers for the methylated PTEN were forward,
5′-GTTTTTTGGGTTTTTGAAATT TAAC-3′ and reverse,
5′-AACCTACTATTATATCGCCAA CGTA-3′, yielding a 166-bp product
(Fig. 1). The 25 μl of PCR mixture
contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.25 mM
MgCl2, 100 μM dNTP, 0.6 μM of each primer, 1 unit of Taq
DNA polymerase (HotStarTaq; Takara Bio, Inc.) and bisulfite
modified DNA (25 ng) (20). The PCR
conditions consisted of a denaturing step at 95°C for 5 min and 35
cycles of 94°C for 30 sec, 60°C (for the detection of the
unmethylated PTEN gene) or 62°C (for the detection of the
methylated PTEN gene) for 30 sec and 72°C for 60 sec, followed by a
final extension at 72°C for 10 min. Each PCR product (20 μl) was
directly loaded onto a 2% agarose gel, stained with ethidium
bromide and visualized under UV illumination. The DNA from the
GenoMethtm Universal Methylated DNA Standard (Zymo Research,
Irvine, CA, USA)was used as a positive control for the methylated
alleles. Water and unmodified DNA were used as negative controls
for every batch reaction. The PCR for all the samples that
demonstrated methylation for the individual genes was repeated to
confirm these results.
Immunohistochemical analysis
Phosphate-buffered saline (PBS) was used instead of
the primary antibody for the blank control and normal non-immunized
rabbit serum was used instead of the primary antibody for the
negative control. Following the manufacturer’s instructions
provided with the immunohistochemistry staining reagent kit
(Boster, Wuhan, China), the deparaffinized tissues were cut into
5-μm thick sections, washed 3 times (5 min each time) with PBS,
incubated at room temperature in 3% H2O2 to
eliminate the endogenous peroxidase activity and subsequently
washed an additional 3 times (3 min each time) with PBS. The
antigens were prepared in a microwave (citrate buffer, pH 6.0),
naturally refrigerated to room temperature, washed 3 times (3 min
each time) with PBS, incubated at room temperature with normal
non-immunized serum solution for 15 min to indicate the
non-specific sites, incubated at room temperature with the primary
antibody solution and horseradish peroxidase-tagged streptavidin
for 15 min and finally washed 3 times (3 min each time) with PBS.
The Drosophila Anion Exchanger (DAE) stain was rinsed with
PBS for 3 min, counterstained with hematoxylin for 1 min, rinsed
with tap water for 2 min, differentiated with 1% hydrochloric
ethanol, rinsed with tap water for 5 min, dehydrated with gradient
alcohol and transparentized with dimethylbenzene. The sections were
coated with neutral balata. PTEN-positive stained slides from
patients with cervicitis were used as the positive control in each
set of experiments. For the negative control, the same procedure
was followed with the exception of the primary antibody. The
staining was graded as either negative (−) or positive (+). The
specimens in which ≥10% of the neoplastic cell cytosols showed a
positive immunoreactivity were considered to be immunoreactive
(Fig. 2). The specificity of the
PTEN antibody (OriGene, Rockville, MD, USA) is an immunoglobulin G1
mouse monoclonal antibody against human PTEN.
Statistical analysis
The χ2 test was used to determine the
correlation of the promoter methylation and the PTEN gene
expression with the clinicopathological parameters. The association
between the promoter methylation with the PTEN gene expression was
also analyzed using the χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Mutational analysis
A total of 102 cases of cervical cancer were
studied. The PTEN novel mutations were identified in one of 102
(1%) tumor specimens. The SSCP analysis showed an abnormal band on
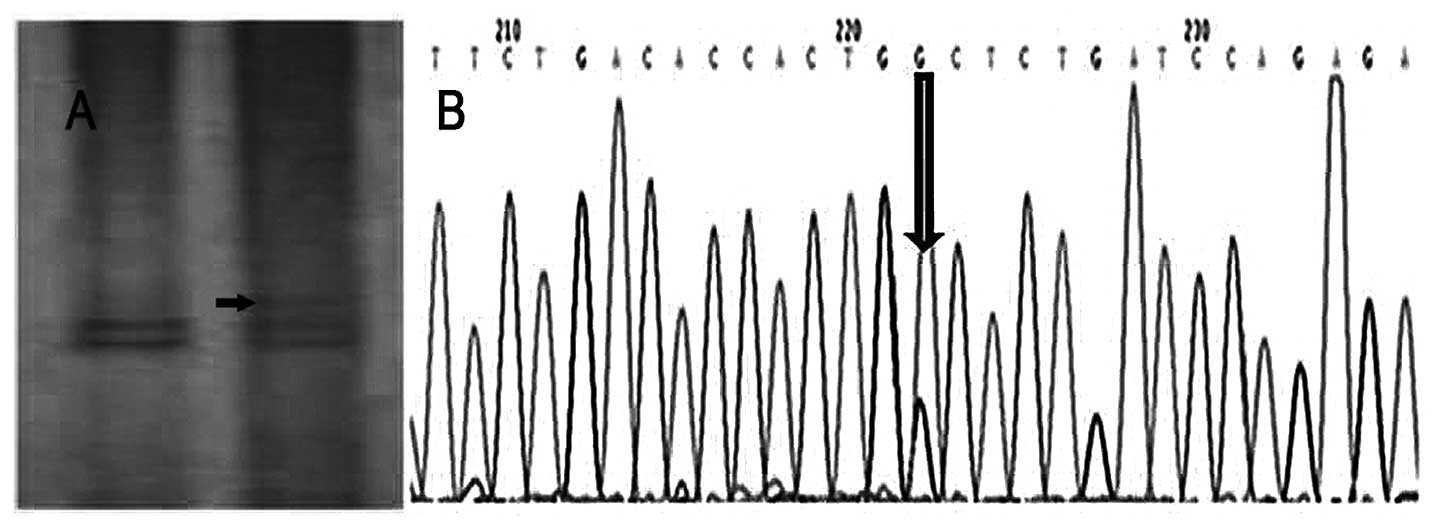
exon 9 of the PTEN gene (Fig. 3A).
Direct sequencing analysis showed the mutation site in exon 9 was
found at codon 380 (CAC-GGC), resulting in an aspartic acid to
glycine substitution (Fig. 3B). No
mutation was found in exons 1 to 8. The clinical parameters of the
mutated patient were evaluated. This patient was 63 years old and
was found to be present in clinical stage III and pathological
grade 2.
Promoter methylation of the PTEN
gene
The PTEN promoter methylation was analyzed in tumor
samples from 102 cervical cancer patients. Among the tested cases,
the PTEN promoter methylation was detected in 63 of 102 (62%) cases
in the tumor DNA. Table II shows
the association between methylation and the clinicopathological
parameters. Among the patients examined for PTEN methylation, 51%
(23 of 45) of cases were methylated in clinical stage I–II and 70%
(40 of 57) in stages III–IV. The correlation between PTEN
methylation and clinical stages was found to be statistically
significant (P=0.039). Furthermore, the study showed that PTEN
methylation was not significantly correlated with the histology
type and age of the patients, but was significantly correlated with
the carcinoma size, lymph node metastasis and tumor grade
(P<0.05).
 | Table IIAssociation between promoter
methylation of the PTEN gene with the clinicopathological features
in cervical cancer patients. |
Table II
Association between promoter
methylation of the PTEN gene with the clinicopathological features
in cervical cancer patients.
| Characteristics | No. of cases tested
(n=102) | Methylation of PTEN
present (%) | P-value |
|---|
| Age, years |
| ≤44 | 44 | 27/44 (61) | 0.552 |
| >44 | 58 | 36/58 (62) | |
| FIGO stage |
| I–II | 45 | 23/45 (51) | 0.039 |
| III–IV | 57 | 40/57 (70) | |
| Tumor grade |
| G1 | 58 | 30/58 (52) | 0.014 |
| G2–G3 | 44 | 33/44 (75) | |
| Tumor size, cm |
| <4 | 63 | 34/63 (54) | 0.031 |
| ≥4 | 39 | 29/39 (74) | |
| Histology type |
| SCC | 78 | 47/78 (60) | 0.376 |
| AD | 24 | 16/24 (67) | |
| Lymph node
metastasis |
| With | 38 | 32/38 (84) | 0.001 |
| Without | 64 | 31/64 (48) | |
PTEN protein expression in patients with
cervical cancer
The immunohistochemical analysis of the PTEN gene
was performed in 102 cases of cervical carcinoma and 15 samples of
cervicitis (inflammatory lesion) as a control. Nuclear PTEN
expression was detected in all the 15 samples of cervicitis and
63/102 (62%) cases of cervical cancer (Table III). A decreased PTEN expression
was observed with the increase in clinical stage (40 and 59% in
stages I–II and III–IV, respectively) and a statistically
significant correlation was also observed between the loss of PTEN
expression and lymph node metastasis. The lymph node metastasis
group showed more expressional loss when compared to the specimens
belonging to the without lymph node metastasis group (P=0.001).
 | Table IIIAssociation between the expression of
the PTEN protein and the clinicopathological features in cervical
cancer. |
Table III
Association between the expression of
the PTEN protein and the clinicopathological features in cervical
cancer.
|
Characteristics | No. of cases tested
(n=102) | Presence of PTEN
expression (%) | P-value |
|---|
| Age, years |
| ≤44 | 44 | 22/44 (50) | 0.511 |
| >44 | 58 | 28/58 (48) | |
| FIGO stage |
| I–II | 45 | 27/45 (60) | 0.038 |
| III–IV | 57 | 23/57 (40) | |
| Tumor grade |
| G1 | 58 | 32/58 (55) | 0.110 |
| G2–G3 | 44 | 18/44 (41) | |
| Tumor size, cm |
| <4 | 63 | 34/63 (54) | 0.143 |
| ≥4 | 39 | 16/39 (41) | |
| Histology type |
| SCC | 78 | 38/78 (49) | 0.549 |
| AD | 24 | 12/24 (50) | |
| Lymph node
metastasis |
| With | 38 | 10/38 (26) | 0.001 |
| Without | 64 | 40/64 (63) | |
Correlation of PTEN expression with
promoter methylation
The correlation of the promoter methylation with the
PTEN gene expression was determined in 102 cases of cervical
cancer. Among 102 cases, 63 were found to be methylated, whereas 37
were identified as unmethylated. The loss of PTEN expression was
observed in 24 of the 63 methylation-positive cases, whereas only
15 of the 39 methylation-negative cases were observed as having a
loss of PTEN expression. The association was found to be
statistically significant, P=0.042 (Table IV).
 | Table IVCorrelation of promoter methylation
with PTEN expression in cervical cancer. |
Table IV
Correlation of promoter methylation
with PTEN expression in cervical cancer.
| PTEN expression
methylation | PTEN promoter |
|---|
|
|---|
| Positive | Negative | P-value |
|---|
| Positive | 26 | 37 | 0.042 |
| Negative | 24 | 15 | |
Discussion
PTEN, a tumor-suppressing gene, is involved in
cellular differentiation, reproduction and apoptosis, as well as
cellular adhesion and mobility. The loss or downregulation of PTEN
plays an important role in the multiple steps of tumorigenesis and
progression of malignancies, and mutations and deletions of this
gene have been described in a wide range of human cancers. The
incidence is 30–50% in endometrial cancers, 25% in glioblastomas,
21% in ovarian cancer, 13% in prostate cancer and <5% in breast
and thyroid cancer (11,19–21).
The mutation of PTEN in cervical carcinoma does not appear to be
frequent. Yaginuma et al (22) reported that one in 43 cervical
cancers (2%) had a PTEN mutation in a Japanese population. Rizvi
et al (23) reported that
three of 135 cervical cancers (2%) had a PTEN mutation in an Indian
population. However, in a Chinese population, Cheung et al
(24) reported 0 of 60 cervical
cancers had a PTEN mutation. In the present study, entire coding
regions of the PTEN gene were screened for the mutation by PCR-SSCP
in 102 cases of cervical cancer. The results showed an A to G point
mutation on exon 9 of the PTEN gene in one case only (case number
36), and this mutation lead to a missense mutation on codon 380,
which caused a transition of aspartic acid into glycine. The exact
function of this mutation is still unknown, and functional analysis
is required to investigate this. The rate of the PTEN gene mutation
is 1% in the patients with cervical cancer from a Chinese
population. It was also reported that PTEN expression was
frequently diminished at the transcriptional level. DNA
methylation, transcriptional repression and microRNA-directed mRNA
degradation and translational disruption appear to reduce PTEN
expression in certain cancers (25,26).
Therefore, it may be concluded that the PTEN mutation is not common
in Chinese cervical cancer.
Recent studies have also shown that the CpG islands
of the PTEN promoter are methylated in various human malignancies
including cervical cancer, but the PTEN methylation data were
different. In a study by Yang et al (18), PTEN methylation was observed in 20
of 127 (16%) cervical cancers, whereas it was reported in 36 of 62
(58%) squamous cell cervical cancers in the study by Cheung et
al (24). There was no PTEN
expression observed in three of 10 and 0 of 10 PTEN
methylation-negative and -positive cases, respectively. The
preliminary data from the study by Eijsink et al (27) showed the presence of PTEN gene
promoter methylation in four of 19 (21%) cases. No PTEN
immunostaining was observed in these four PTEN methylation-positive
cases. Rizvi et al (23)
reported that in the PTEN gene, there was methylation in 61%
(83/135) of specimens and a loss of PTEN expression in 41% (34/83)
of methylation-positive cases, whereas in the 52
methylation-negative cases only 25% (13/52) were observed as
immunostaining-negative, which was statistically significant
(P=0.05). In the present study, PTEN promoter methylation was
detected in 63 of 102 (62%) cases in the tumor DNA. PTEN
methylation was significantly correlated with the FIGO stage,
carcinoma size, lymph node metastasis and tumor grade (P<0.05).
Any discrepancies between the data of the PTEN methylation from the
high and the low rates may be due to the number of evaluated
patients and/or the samples with/without the radiotherapy and/or
chemotherapy. The patients in the study by Yang et al
(18) received radiotherapy and/or
chemotherapy, and the study by Eijsink et al (27) only analyzed 19 cases.
Furthermore, the present study showed that the PTEN
protein expression in the 102 cervical cancer tissue samples was
significantly correlated with the FIGO stage and lymph node
metastasis (P<0.05). The Lu et al (28) study showed that the level of PTEN
expression in the metastatic pelvic lymph node group was
significantly lower compared to the non-metastatic group
(P<0.01). The Eijsink et al (27) study indicated that a loss of PTEN
expression frequently occurs in early-stage cervical cancer and
this is associated with pelvic lymph node metastasis (P=0.019), but
not to the survival rate. These experimental results suggest that
there is a role for the loss of PTEN in determining the metastatic
potential of tumors.
Therefore, a correlation between the occurrence of
PTEN promoter methylation with its reduced expression was also
formulated. Promoter methylation of PTEN was significantly
correlated with the loss of PTEN expression and showed a
significant association (P=0.042). These results indicate that a
reduced PTEN expression may be due to PTEN promoter methylation.
Promoter methylation of PTEN presents a major alternative mechanism
of gene silencing.
In conclusion, abnormal promoter methylation of the
PTEN gene is usually found in cervical cancer and is associated
with tumor differentiation, infiltrating depth, lymph node
metastasis and FIGO staging. PTEN may play an important role in the
occurrence and development of cervical cancer, and the PTEN protein
expression phenotype can be considered as an indicator for the
pathophysiological behavior of cervical cancer.
Acknowledgements
The authors would like to thank the personnel at the
Changzhou Tumor Hospital (Changzhou, China) who were involved in
the present study. The study was partly supported by the Natural
Science Foundation of China (grant nos. 81071799 and 81372212), the
Natural Science Foundation of Jiangsu (grant nos. BK2011251 and
BL2013012), the Health Talents Project for Jiangsu, China (grant
nos. LJ201157, RC2011038 and BRA2011038), the Research of Health
Department in Jiangsu (grant no. Z201221), the Talents Project
(‘831’ and health) of Changzhou Municipality, the Science and
Technology Planning Project of Changzhou Health Bureau, Jiangsu
(grant nos. QN201106 and ZD201203), the 333 Talents Training
Project of Jiangsu Province, the Key Medical Innovation Talents
Training Project of Changzhou, Jiangsu, and the Project of Jiangsu
Province Sanitation Innovation Team.
References
|
1
|
Ferlay J, Shin HR, Bray F, et al:
Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int
J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Walboomers JM, Jacobs MV, Manos MM, et al:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Noordhuis MG, Eijsink JJ, Ten HK, et al:
Expression of epidermal growth factor receptor (EGFR) and activated
EGFR predict poor response to (chemo) radiation and survival in
cervical cancer. Clin Cancer Res. 15:7389–7397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woelber L, Kress K, Kersten JF, et al:
Carbonic anhydrase IX in tumor tissue and sera of patients with
primary cervical cancer. BMC Cancer. 11:122011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hellberg D, Tot T and Stendahl U: Pitfalls
in immunohistochemical validation of tumor marker expression -
exemplified in invasive cancer of the uterine cervix. Gynecol
Oncol. 112:235–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khunamornpong S, Settakorn J, Sukpan K, et
al: Cyclooxygenase-2 expression in squamous cell carcinoma of the
uterine cervix is associated with lymph node metastasis. Gynecol
Oncol. 112:241–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lima EM, Araújo JJ, Harada ML, et al:
Molecular study of the tumour suppressor gene PTEN in gastric
adenocarcinoma in Brazil. Clin Exp Med. 5:129–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oki E, Kakeji Y, Baba H, et al: Impact of
loss of heterozygosity of encoding phosphate and tensin homolog on
the prognosis of gastric cancer. J Gastroenterol Hepatol.
21:814–818. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stiles BL: Phosphatase and tensin
homologue deleted on chromosome 10: extending its PTENtacles. Int J
Biochem Cell Biol. 41:757–761. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Raftopoulou M, Etienne-Manneville S, Self
A, et al: Regulation of cell migration by the C2 domain of the
tumor suppressor PTEN. Science. 303:1179–1181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li J, Yen C, Liaw D, et al: PTEN, a
putative protein tyrosine phosphatase gene mutated in human brain,
breast and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo CY, Xu XF, Wu JY and Liu SF:
PCR-SSCP-DNA sequencing method in detecting PTEN gene mutation and
its significance in human gastric cancer. World J Gastroenterol.
14:3804–3811. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marsit CJ, Zheng S, Aldape K, et al: PTEN
expression in non-small-cell lung cancer: evaluating its relation
to tumor characteristics, allelic loss, and epigenetic alteration.
Hum Pathol. 36:768–776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Teng Y, Sun AN, Pan XC, et al: Synergistic
function of Smad4 and PTEN in suppressing forestomach squamous cell
carcinoma in the mouse. Cancer Res. 66:6972–6981. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Perrone F, Lampis A, Orsenigo M, et al:
PI3KCA/PTEN deregulation contributes to impaired responses to
cetuximab in metastatic colorectal cancer patients. Ann Oncol.
20:84–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang YH, Lee HS and Kim WH: Promoter
methylation and silencing of PTEN in gastric carcinoma. Lab Invest.
82:285–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Soria JC, Lee HY, Lee JI, et al: Lack of
PTEN expression in non-small cell lung cancer could be related to
promoter methylation. Clin Cancer Res. 8:1178–1184. 2002.PubMed/NCBI
|
|
18
|
Yang HJ, Liu VW, Wang Y, et al:
Differential DNA methylation profiles in gynecological cancers and
correlation with clinico-pathological data. BMC Cancer. 6:2122006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang YA, Fan LF, Jiang CQ, et al:
Expression and significance of PTEN, hypoxia-inducible factor-1
alpha in colorectal adenoma and adenocarcinoma. World J
Gastroenterol. 9:491–494. 2003.PubMed/NCBI
|
|
20
|
Schöndorf T, Ebert MP, Hoffmann J, et al:
Hypermethylation of the PTEN gene in ovarian cancer cell lines.
Cancer Lett. 207:215–220. 2004.PubMed/NCBI
|
|
21
|
Steck PA, Pershouse MA, Jasser SA, et al:
Identification of a candidate tumour suppressor gene, MMAC1, at
chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yaginuma Y, Yamashita T, Ishiya T, et al:
Abnormal structure and expression of PTEN/MMAC1 gene in human
uterine cancers. Mol Carcinog. 27:110–116. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rizvi MM, Alam MS, Ali A, et al: Aberrant
promoter methylation and inactivation of PTEN gene in cervical
carcinoma from Indian population. J Cancer Res Clin Oncol.
137:1255–1262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheung TH, Lo KW, Yim SF, et al:
Epigenetic and genetic alternation of PTEN in cervical neoplasm.
Gynecol Oncol. 93:621–627. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wiencke JK, Zheng S, Jelluma N, et al:
Methylation of the PTEN promoter defines low-grade gliomas and
secondary glioblastoma. Neuro Oncol. 9:271–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eijsink JJ, Noordhuis MG, ten Hoor KA, et
al: The epidermal growth factor receptor pathway in relation to
pelvic lymph node metastasis and survival in early-stage cervical
cancer. Hum Pathol. 41:1735–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu D, Qian J, Yin X, et al: Expression of
PTEN and survivin in cervical cancer: promising biological markers
for early diagnosis and prognostic evaluation. Br J Biomed Sci.
69:143–146. 2012.PubMed/NCBI
|