Introduction
The prevalence of type 2 diabetes mellitus (DM) is
increasing rapidly worldwide, with >170 million individuals
currently affected (1,2) and 439 million adults (7.7% of all adults)
predicted to be affected by 2030 (2).
The major site of this emerging epidemic is expected to be Asia,
mainly as a result of changes in nutrition and other lifestyle
factors (3). Given that type 2 DM
increases the risk for cardiovascular disease and long-term
mortality, the health care burden imposed by this condition is a
matter of urgent concern (4,5). Aggressive strategies for disease
prevention and early detection will be key to tackling this global
issue. Approximately 95% of patients with DM have type 2 DM, with
characteristics that range from insulin resistance with relatively
minor insulin deficiency to insulin deficiency with relatively
minor insulin resistance (6). The
several mechanisms that have been suggested for the pathogenesis of
type 2 DM include an increase in the production of nonesterified
fatty acids, inflammatory cytokines or adipokines, and dysfunction
of mitochondria for insulin resistance and glucotoxicity,
lipotoxicity and amyloid formation for β-cell dysfunction (1). Although a sedentary lifestyle and
overeating appear to be triggering factors, genetic factors are
also indicated in the pathogenesis of type 2 DM, as a positive
family history is associated with a 2.4-fold increase in the risk
(1,7).
Previous genome-wide association studies (GWASs)
have indicated numerous loci and genes in the predisposition to
type 2 DM in various ethnic groups (8–17). Although
KCNQ1 (13,14) and UBE2E2 (15) were identified as susceptibility genes
for type 2 DM in Japanese individuals, the genes that contribute to
genetic susceptibility to this condition remain to be
identified.
Our previous studies identified nine genes and
chromosomal region 3q28 as susceptibility loci for myocardial
infarction, ischemic stroke or chronic kidney disease in Japanese
individuals by genome-wide (18–20) or
candidate gene (21–23) association studies. As type 2 DM is an
important risk factor for these diseases (24–26), we
hypothesized that certain single-nucleotide polymorphisms (SNPs) at
these 10 loci may contribute to the genetic susceptibility by
affecting the susceptibility to type 2 DM. The present study
examined the possible association of 13 SNPs at the 10 loci with
the prevalence of type 2 DM in community-dwelling Japanese
individuals.
Materials and methods
Study population
Study subjects comprised 6,027 community-dwelling
individuals (797 subjects with type 2 DM and 5,230 controls) who
were recruited to a population-based cohort study (Inabe Health and
Longevity Study) in Inabe (Mie, Japan). The Inabe Health and
Longevity Study is a longitudinal genetic epidemiological study of
atherosclerotic, cardiovascular and metabolic diseases (27–33).
Detailed methods for recruitment of study subjects and collection
of medical examination data were described previously (27).
Individuals with DM were defined as those with a
fasting plasma glucose concentration of ≥126 mg/dl (6.93 mmol/l),
with a blood glycosylated hemoglobin (hemoglobin A1c)
content of ≥6.5%, or who were taking antidiabetic medication. Type
2 DM was defined according to the criteria accepted by the World
Health Organization and described previously (6,34).
Individuals with type 1 DM, maturity-onset diabetes of the young,
DM associated with mitochondrial diseases or single-gene disorders,
pancreatic diseases, including severe pancreatitis and pancreatic
tumors, or other metabolic or endocrinological diseases were
excluded from the study. Individuals taking drugs that cause
secondary DM were also excluded. Control individuals had a fasting
plasma glucose level of <110 mg/dl (6.05 mmol/l), a blood
hemoglobin A1c content of <6.2% and no history of DM
or of taking antidiabetic medication.
The study protocol complied with the Declaration of
Helsinki and was approved by the Committees on the Ethics of Human
Research of Mie University Graduate School of Medicine and Inabe
General Hospital (Mie, Japan). Written informed consent was
obtained from all the subjects.
Selection and genotyping of
polymorphisms
The 13 SNPs examined in the present study were
selected from our previous genome-wide (18–20) or
candidate gene (21–23) association studies and were described
previously (27). Wild-type
(ancestral) and variant alleles of the SNPs were determined from
the SNP database (dbSNP, National Center for Biotechnology
Information, Bethesda, MD, USA; http://www.ncbi.nlm.nih.gov/SNP).
Venous blood (5 ml) was collected into tubes
containing 50 mmol/l ethylenediaminetetraacetic acid (disodium
salt), peripheral blood leukocytes were isolated and genomic DNA
was extracted from these cells with a DNA extraction kit (SMITEST
EX-R&D; Medical and Biological Laboratories, Co., Ltd., Nagoya,
Japan). Genotypes of the 13 SNPs were determined at G&G Science
Co., Ltd., (Fukushima, Japan) by a method that combines the
polymerase chain reaction and sequence-specific oligonucleotide
probes with suspension array technology (Luminex Corp., Austin, TX,
USA). Primers, probes and other conditions for genotyping of SNPs
examined in the present study were described previously (27), as was the detailed genotyping
methodology (35).
Statistical analysis
Quantitative data were compared between subjects
with type 2 DM and controls using the unpaired Student's t-test.
Categorical data were compared with the χ2 test. The
associations of 13 SNPs to the prevalence of type 2 DM, to fasting
plasma glucose level or blood hemoglobin A1c content
were examined in a 5-year longitudinal cohort study. Longitudinal
changes in the prevalence of type 2 DM were compared between the
two groups (dominant or recessive genetic model) by a generalized
estimating equation (36) and with
adjustment for age, gender and body mass index (BMI). Longitudinal
changes in fasting plasma glucose level or blood hemoglobin
A1c content in all the individuals or in individuals not
taking antidiabetic medication were compared between the two groups
(dominant or recessive model) in a generalized linear mixed-effect
model (37) with adjustment for age,
gender and BMI. Age-related changes in the prevalence of type 2 DM
or in fasting plasma glucose level or blood hemoglobin
A1c content were estimated with quadratic curves
controlling for the observation year. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analysis was performed with R software version 3-0-2 (The R Project
for Statistical Computing) and JMP Genomics version 6.0 (SAS
Institute, Inc., Cary, NC, USA).
Results
Subject characteristics
Characteristics of subjects with type 2 DM and
controls in the cross-sectional analysis in March 2014 are shown in
Table I. Age, the frequency of males
and BMI were significantly greater in subjects with type 2 DM
compared to the controls.
 | Table I.Characteristics of the subjects with
type 2 diabetes mellitus and controls: Cross-sectional analysis in
March 2014. |
Table I.
Characteristics of the subjects with
type 2 diabetes mellitus and controls: Cross-sectional analysis in
March 2014.
| Parameter | Diabetes mellitus
(n) | Controls (n) | P-value |
|---|
| No. of
subjects | 797 | 5230 |
|
| Age, years | 61.9±10.5
(797) | 53.0±12.9
(5230) | <0.0001 |
| Gender, %
(male/female) | 70.5/29.5
(797) | 53.3/46.7
(5230) | <0.0001 |
| Height, cm | 162.6±9.7
(760) | 162.5±9.1
(5194) | 0.7605 |
| Weight, kg | 64.6±13.5
(758) | 60.4±11.8
(5194) | <0.0001 |
| Body mass index,
kg/m2 | 24.3±3.9 (758) | 22.8±3.3
(5194) | <0.0001 |
| Waist
circumference, cm | 84.9±9.9 (683) | 79.8±9.0
(4922) | <0.0001 |
| Alcohol drinking,
% | 50.2 (797) | 48.0 (5230) | 0.2437 |
| Current or former
smoking, % | 55.5 (797) | 44.2 (5230) | <0.0001 |
| Systolic blood
pressure, mmHg | 127±18 (753) | 120±16 (5192) | <0.0001 |
| Diastolic blood
pressure, mmHg | 77±12 (753) | 74±12 (5192) | <0.0001 |
| Mean blood
pressure, mmHg | 94±13 (753) | 89±12 (5192) | <0.0001 |
| Ocular tension,
right, mmHg | 14.1±3.2 (246) | 13.4±2.9
(1815) | 0.0005 |
| Functional vital
capacity, l | 3.14±0.78
(255) | 3.32±0.81
(1988) | 0.0009 |
| FEV1% | 80.4±6.3 (255) | 81.4±6.6
(1988) | 0.0287 |
| Serum albumin,
g/l | 44.1±3.6 (613) | 44.7±2.5
(3599) | <0.0001 |
| Serum total
cholesterol, mmol/l | 5.10±1.00
(784) | 5.23±0.87
(5166) | 0.0001 |
| Serum
triglycerides, mmol/l | 1.49±1.06
(772) | 1.23±0.82
(5164) | <0.0001 |
| Serum
HDL-cholesterol, mmol/l | 1.51±0.42
(771) | 1.68±0.45
(5163) | <0.0001 |
| Serum
LDL-cholesterol, mmol/l | 3.13±0.87
(770) | 3.18±0.79
(5162) | 0.1294 |
| Fasting plasma
glucose, mg/dl | 132.4±40.3
(789) | 95.8±8.6
(5167) | <0.0001 |
| Blood hemoglobin
A1c, % | 6.65±1.27
(621) | 5.54±0.33
(3842) | <0.0001 |
| Blood urea
nitrogen, mmol/l | 6.27±3.48
(612) | 5.03±1.54
(3489) | <0.0001 |
| Serum creatinine,
µmol/l | 109.5±182.9
(767) | 68.3±45.3
(4809) | <0.0001 |
| eGFR, ml
min−1 1.73 m−2 | 71.2±23.6
(767) | 77.5±15.2
(4809) | <0.0001 |
| Serum uric acid,
µmol/l | 340±84 (759) | 324±86 (4772) | <0.0001 |
| Serum C-reactive
protein, µg/l | 2515±14479
(295) | 981±3758
(1818) | 0.0001 |
| White blood cells,
103/µl | 5.90±2.19
(554) | 5.31±1.57
(4053) | <0.0001 |
| Red blood cells,
104/µl | 438±50 (556) | 437±44 (4067) | 0.4542 |
| Hemoglobin,
g/l | 139±18 (556) | 138±15 (4067) | 0.0549 |
| Hematocrit, % | 40.6±4.8 (555) | 40.2±4.2
(4063) | 0.0656 |
| Platelets,
104/µl | 21.0±5.9 (551) | 22.5±5.3
(4017) | <0.0001 |
Associations with type 2 DM
The associations of the 13 SNPs to the prevalence of
type 2 DM were analyzed with a generalized estimating equation and
with adjustment for age, gender and BMI (Table II). The rs2116519 (C→T) SNP of the
FAM78B gene (recessive model), as well as rs2074379 (G→A,
dominant model) and rs2074388 (A→G, dominant model) of ALPK1
were significantly (P<0.05) associated with the prevalence of
type 2 DM.
 | Table II.Associations of polymorphisms with
type 2 diabetes mellitus analyzed for 5-year longitudinal data with
a generalized estimating equation. |
Table II.
Associations of polymorphisms with
type 2 diabetes mellitus analyzed for 5-year longitudinal data with
a generalized estimating equation.
| Gene or locus | SNP | Genotype | Diabetes mellitus,
n (%) | Control, n (%) | P-value
(dominant)a | P-value
(recessive)b |
|---|
| FAM78B | rs2116519
(C→T) | TT | 553 (29.0) | 5783 (31.0) | 0.0648 | 0.0188c |
|
|
| TC | 950 (49.8) | 9438 (50.5) |
|
|
|
|
| CC | 403 (21.1) | 3459 (18.5) |
|
|
| 3q28 | rs9846911
(A→G) | AA | 1642 (86.1) | 16272 (87.1) | 0.7924 | 0.9172 |
|
|
| AG | 251 (13.2) | 2304 (12.3) |
|
|
|
|
| GG | 13 (0.7) | 104 (0.6) |
|
|
| ALPK1 | rs2074379
(G→A) | AA | 807 (42.3) | 8680 (46.5) | 0.0121c | 0.2027 |
|
|
| AG | 901 (47.3) | 8180 (43.8) |
|
|
|
|
| GG | 198 (10.4) | 1820 (9.7) |
|
|
| ALPK1 | rs2074380
(G→A) | GG | 1613 (84.6) | 15797 (84.6) | 0.6579 | 0.3014 |
|
|
| GA | 287 (15.1) | 2736 (14.6) |
|
|
|
|
| AA | 6 (0.3) | 147 (0.8) |
|
|
| ALPK1 | rs2074381
(A→G) | AA | 1646 (86.4) | 15937 (85.3) | 0.4558 | 0.1330 |
|
|
| AG | 258 (13.5) | 2622 (14.0) |
|
|
|
|
| GG | 2 (0.1) | 121 (0.6) |
|
|
| ALPK1 | rs2074388
(A→G) | AA | 799 (41.9) | 8687 (46.5) | 0.0053c | 0.1492 |
|
|
| AG | 906 (47.5) | 8169 (43.7) |
|
|
|
|
| GG | 201 (10.5) | 1824 (9.8) |
|
|
| BTN2A1 | rs6929846
(T→C) | CC | 1507 (79.1) | 14566 (78.0) | 0.6322 | 0.9904 |
|
|
| CT | 373 (19.6) | 3850 (20.6) |
|
|
|
|
| TT | 26 (1.4) | 264 (1.4) |
|
|
| THBS2 | rs8089 (T→G) | TT | 1559 (81.8) | 15393 (82.4) | 0.7768 | 0.4105 |
|
|
| TG | 329 (17.3) | 3120 (16.7) |
|
|
|
|
| GG | 18 (0.9) | 167 (0.9) |
|
|
| PDX1 | rs146021107
(G→-) | GG | 540 (28.3) | 5303 (28.4) | 0.9381 | 0.6936 |
|
|
| G– | 924 (48.5) | 9284 (49.7) |
|
|
|
|
| −− | 442 (23.2) | 4093 (21.9) |
|
|
| F7 | rs6046 (G→A) | GG | 1665 (87.4) | 16285 (87.2) | 0.6075 | 0.1153 |
|
|
| GA | 234 (12.3) | 2313 (12.4) |
|
|
|
|
| AA | 7 (0.4) | 82 (0.4) |
|
|
| LLGL2 | rs1671021
(G→A) | AA | 1393 (73.1) | 13813 (73.9) | 0.7072 | 0.5812 |
|
|
| AG | 487 (25.6) | 4490 (24.0) |
|
|
|
|
| GG | 26 (1.4) | 377 (2.0) |
|
|
| ILF3 | rs2569512
(G→A) | GG | 834 (43.8) | 8200 (43.9) | 0.8400 | 0.4642 |
|
|
| GA | 878 (46.1) | 8442 (45.2) |
|
|
|
|
| AA | 194 (10.2) | 2038 (10.9) |
|
|
| CELSR1 | rs6007897
(C→T) | TT | 1838 (96.4) | 18151 (97.2) | 0.1157 | ND |
|
|
| TC | 68 (3.6) | 529 (2.8) |
|
|
|
|
| CC | 0 (0) | 0 (0) |
|
|
The associations between the prevalence of type 2 DM
and age analyzed longitudinally with a generalized estimating
equation according to SNP genotype are shown in Fig. 1. The prevalence of type 2 DM was
greater in subjects with the CC genotype of rs2116519 of
FAM78B compared to the combined group of subjects with the
TT or TC genotypes from 40 to 90 years of age
(Fig. 1A), in the combined group of
subjects with the AG or GG genotypes of rs2074379 of
ALPK1 compared to those with the AA genotype
(Fig. 1B) and in the combined group of
subjects with the AG or GG genotypes of rs2074388 of
ALPK1 compared to those with the AA genotype
(Fig. 1C).
As three SNPs were significantly associated with
type 2 DM, the associations of these SNPs to fasting plasma glucose
level or blood hemoglobin A1c content in all the
individuals or individuals not taking antidiabetic medication were
analyzed with a generalized linear mixed-effect model and with
adjustment for age, gender and BMI (Table III). The rs2116519 SNP of
FAM78B, as well as rs2074379 and rs2074388 of ALPK1
were significantly (P<0.05) associated with fasting plasma
glucose level and blood hemoglobin A1c content in a
dominant model among all the individuals. Among individuals not
taking antidiabetic medication, rs2116519 of FAM78B was
significantly associated with blood hemoglobin A1c
content in a dominant model, whereas rs2074379 and rs2074388 of
ALPK1 were significantly associated with fasting plasma
glucose level and blood hemoglobin A1c content in a
dominant model.
 | Table III.Associations of polymorphisms to
fasting plasma glucose level or blood hemoglobin A1c
content in all individuals or individuals not taking antidiabetic
medication, analyzed for 5-year longitudinal data with a
generalized linear mixed-effect model. |
Table III.
Associations of polymorphisms to
fasting plasma glucose level or blood hemoglobin A1c
content in all individuals or individuals not taking antidiabetic
medication, analyzed for 5-year longitudinal data with a
generalized linear mixed-effect model.
| Gene (SNP) | Dominant
modela | P-value | Recessive
modela | P-value |
|---|
| All
individuals |
|
|
|
|
|
|
|
FAM78B (rs2116519,
C→T) | TT
(6336) | TC +
CC (14250) |
| TT +
TC (16724) | CC
(3862) |
|
Fasting plasma
glucose, mg/dl | 99.9±16.4 | 100.5±18.3 | 0.0352b | 100.3±17.5 | 100.5±19.0 | 0.2251 |
|
Blood hemoglobin
A1c, % | 5.69±0.59 | 5.71±0.65 | 0.0065b | 5.70±0.63 | 5.70±0.66 | 0.4079 |
|
ALPK1 (rs2074379,
G→A) | AA
(9484) | AG +
GG (11099) |
| AA +
AG (18568) | GG
(2018) |
|
Fasting plasma
glucose, mg/dl | 99.8±15.7 | 100.8±19.4 | 0.0017b | 100.3±17.6 | 101.0±19.0 | 0.5509 |
|
Blood hemoglobin
A1c, % | 5.68±0.57 | 5.72±0.69 | 0.0090b | 5.70±0.62 | 5.74±0.73 | 0.0502 |
|
ALPK1 (rs2074388,
A→G) | AA
(9486) | AG +
GG (11100) |
| AA +
AG (18561) | GG
(2025) |
|
Fasting plasma
glucose, mg/dl | 99.8±15.6 | 100.8±19.4 | 0.0010b | 100.3±17.6 | 101.2±19.1 | 0.4149 |
|
Blood hemoglobin
A1c, % | 5.68±0.57 | 5.72±0.69 | 0.0079b | 5.70±0.62 | 5.74±0.73 | 0.0417 |
| Individuals without
antidiabetic medication |
|
|
|
|
|
|
|
FAM78B, (rs2116519,
C→T) | TT
(6260) | TC +
CC (14045) |
| TT +
TC (16495) | CC
(3810) |
|
|
Fasting plasma
glucose, mg/dl | 99.6±15.8 | 100.0±17.2 | 0.1087 | 99.8±16.6 | 99.9±17.6 | 0.2482 |
|
Blood hemoglobin
A1c, % | 5.68±0.57 | 5.69±0.61 | 0.0470b | 5.68±0.60 | 5.68±0.62 | 0.3992 |
|
ALPK1, (rs2074379,
G→A) | AA
(9370) | AG +
GG (10935) |
| AA +
AG (18318) | GG
(1987) |
|
|
Fasting plasma
glucose, mg/dl | 99.4±15.0 | 100.2±18.1 | 0.0073b | 99.8±16.6 | 100.5±18.3 | 0.5845 |
|
Blood hemoglobin
A1c, % | 5.66±0.54 | 5.70±0.65 | 0.0142b | 5.68±0.59 | 5.72±0.71 | 0.1134 |
|
ALPK1, (rs2074388,
A→G) | AA
(9369) | AG +
GG (10936) |
| AA +
AG (18311) | GG
(1994) |
|
|
Fasting plasma
glucose, mg/dl | 99.4±15.0 | 100.3±18.2 | 0.0042b | 99.8±16.6 | 100.6±18.3 | 0.4372 |
|
Blood hemoglobin
A1c, % | 5.66±0.54 | 5.70±0.65 | 0.0126b | 5.68±0.59 | 5.72±0.71 | 0.0947 |
The associations between fasting plasma glucose
level and age analyzed longitudinally according to genotype in all
the individuals with a generalized linear mixed-effect model are
shown in Fig. 2. Fasting plasma
glucose level was greater in the combined group of individuals with
the TC or CC genotypes of rs2116519 of FAM78B
compared to those with the TT genotype from 40 to 90 years
of age (Fig. 2A), in the combined
group of individuals with the AG or GG genotypes of
rs2074379 of ALPK1 compared to those with the AA
genotype (Fig. 2B) and in the combined
group of individuals with the AG or GG genotypes of
rs2074388 of ALPK1 compared to those with the AA
genotype (Fig. 2C).
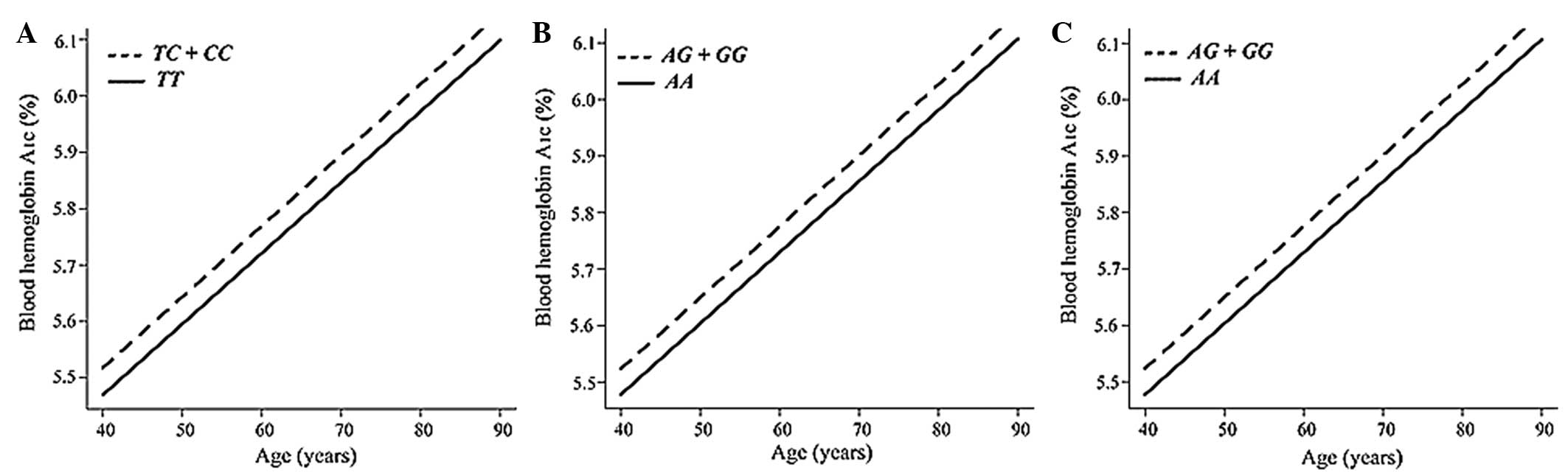
The associations between blood hemoglobin
A1c content and age analyzed longitudinally according to
genotype in all the individuals with a generalized linear
mixed-effect model are shown in Fig.
3. Blood hemoglobin A1c was greater in the combined
group of individuals with the TC or CC genotypes of
rs2116519 of FAM78B compared to those with the TT
genotype from 40 to 90 years of age (Fig.
3A), in the combined group of individuals with the AG or
GG genotypes of rs2074379 of ALPK1 compared to those
with the AA genotype (Fig. 3B)
and in the combined group of individuals with the AG or
GG genotypes of rs2074388 of ALPK1 compared to those
with the AA genotype (Fig.
3C).
Discussion
As genetic factors and interactions between multiple
genes and environmental factors are important in the development of
type 2 DM (1,7), prediction of the risk for type 2 DM on
the basis of genetic variants would be beneficial for personalized
prevention of this condition. In the present study, rs2074379 and
rs2074388 of ALPK1 were significantly associated with the
prevalence of type 2 DM in a longitudinal genetic epidemiological
study, with the minor G allele of each SNP representing a
risk factor for this condition. Our previous study showed that
ALPK1 is a susceptibility locus for chronic kidney disease
in individuals with DM by a GWAS (20). We also observed that genetic variants
of ALPK1 were associated with type 2 DM in a previous
cross-sectional analysis of the Inabe Health and Longevity Study
(28). The present results in the
longitudinal population-based study are consistent with the
previous observations in the cross-sectional study (28) and they validate the association of
genetic variants of ALPK1 with type 2 DM.
ALPK1 functions in apical transport by
phosphorylating myosin 1a in epithelial cells and is indicated in
the regulation of intracellular trafficking processes by
phosphorylation (38). ALPK1 may act
synergistically with monosodium urate monohydrate crystals to
promote the production of proinflammatory cytokines through the
activation of nuclear factor-κB and mitogen-activated protein
kinase (extracellular signal-regulated kinase 1/2 and p38)
signaling in cultured HEK293 cells, suggesting that ALPK1 may
contribute to the inflammatory process associated with the
development of gout (39).
Impaired insulin secretion and increased insulin
resistance are key components of type 2 DM (40). Although the contributions of these
factors to the onset and progression of type 2 DM may differ
between Caucasian and Asian populations, the two factors are
significant for diagnostic and therapeutic strategies targeted to
this disease (41). Previous studies
have shown that proinflammatory cytokines (interleukin-1β and tumor
necrosis factor) detrimentally affect insulin secretion and
resistance (42,43). Additionally, signaling pathways
activated by proinflammatory cytokines, including those mediated by
nuclear factor-κB, have been identified to impair insulin secretion
or to promote insulin resistance (44). As chronic inflammation may play an
important role in the development of type 2 DM, the effects of
rs2074379 and rs2074388 of ALPK1 on the inflammatory process
may account for the association of this gene with type 2 DM.
rs2116519 of FAM78B was also associated with
the prevalence of type 2 DM, as well as to fasting plasma glucose
level and blood hemoglobin A1c content among all the
individuals or to blood hemoglobin A1c content among the
individuals not taking antidiabetic medication. FAM78B is
located at chromosome 1q24.1, a region previously suggested to
harbor a susceptibility locus for type 2 DM (45), although the function of this gene
remains unclear.
There are certain limitations to the present study:
i) As the results were not replicated, validation of these findings
requires their replication with other independent subject panels or
ethnic groups; ii) rs2074379 or rs2074388 are possibly in linkage
disequilibrium with other polymorphisms in the same gene or in
other nearby genes that are responsible for the development of type
2 DM; and iii) the functional relevance of rs2074379 or rs2074388
of ALPK1 to the pathogenesis of type 2 DM has not been
determined.
In conclusion, the present results suggest that
ALPK1 is a susceptibility gene for type 2 DM in
community-dwelling Japanese individuals. Determination of genotypes
for the polymorphisms of ALPK1 may prove informative for
assessment of the genetic risk for type 2 DM in the Japanese
population.
Acknowledgements
The present study was supported by Core Research for
Evolutional Science and Technology of the Japan Science and
Technology Agency (Y.Y. and I.T.) and by a Grant-in-Aid for
Scientific Research from the Ministry of Education, Culture,
Sports, Science and Technology of Japan (grant no. 24590746 to
Y.Y.).
References
|
1
|
Stumvoll M, Goldstein BJ and van Haeften
TW: Type 2 diabetes: Principles of pathogenesis and therapy.
Lancet. 365:1333–1346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaw JE, Sicree RA and Zimmet PZ: Global
estimates of the prevalence of diabetes for 2010 and 2030. Diabetes
Res Clin Pract. 87:4–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan JC, Malik V, Jia W, Kadowaki T,
Yajnik CS, Yoon KH and Hu FB: Diabetes in Asia: Epidemiology, risk
factors, and pathophysiology. JAMA. 301:2129–2140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang W, Dall TM, Halder P, et al: Economic
costs of diabetes in the U.S. in 2012. Diabetes Care. 36:1033–1046.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang P, Zhang X, Brown J, Vistisen D,
Sicree R, Shaw J and Nichols G: Global healthcare expenditure on
diabetes for 2010 and 2030. Diabetes Res Clin Pract. 87:293–301.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Expert Committee on the Diagnosis and
Classification of Diabetes Mellitus: Report of the expert committee
on the diagnosis and classification of diabetes mellitus. Diabetes
Care. 26:(Suppl 1). S5–S20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu FB: Globalization of diabetes: The role
of diet, lifestyle, and genes. Diabetes Care. 34:1249–1257. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wellcome Trust Case Control Consortium:
Genome-wide association study of 14,000 cases of seven common
diseases and 3,000 shared controls. Nature. 447:661–678. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sladek R, Rocheleau G, Rung J, et al: A
genome-wide association study identifies novel risk loci for type 2
diabetes. Nature. 445:881–885. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steinthorsdottir V, Thorleifsson G,
Reynisdottir I, et al: A variant in CDKAL1 influences insulin
response and risk of type 2 diabetes. Nat Genet. 39:770–775. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bouatia-Naji N, Bonnefond A,
Cavalcanti-Proença C, et al: A variant near MTNR1B is associated
with increased fasting plasma glucose levels and type 2 diabetes
risk. Nat Genet. 41:89–94. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Voight BF, Scott LJ, Steinthorsdottir V,
et al: Twelve type 2 diabetes susceptibility loci identified
through large-scale association analysis. Nat Genet. 42:579–589.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yasuda K, Miyake K, Horikawa Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Unoki H, Takahashi A, Kawaguchi T, et al:
SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes
in East Asian and European populations. Nat Genet. 40:1098–1102.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamauchi T, Hara K, Maeda S, et al: A
genome-wide association study in the Japanese population identifies
susceptibility loci for type 2 diabetes at UBE2E2 and
C2CD4A-C2CD4B. Nat Genet. 42:864–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho YS, Chen CH, Hu C, et al:
Meta-analysis of genome-wide association studies identifies eight
new loci for type 2 diabetes in east Asians. Nat Genet. 44:67–72.
2012. View
Article : Google Scholar
|
|
17
|
Kooner JS, Saleheen D, Sim X, et al:
Genome-wide association study in individuals of South Asian
ancestry identifies six new type 2 diabetes susceptibility loci.
Nat Genet. 43:984–989. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamada Y, Nishida T, Ichihara S, et al:
Association of a polymorphism of BTN2A1 with myocardial infarction
in East Asian populations. Atherosclerosis. 215:145–152. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamada Y, Fuku N, Tanaka M, et al:
Identification of CELSR1 as a susceptibility gene for ischemic
stroke in Japanese individuals by a genome-wide association study.
Atherosclerosis. 207:144–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamada Y, Nishida T, Ichihara S, et al:
Identification of chromosome 3q28 and ALPK1 as susceptibility loci
for chronic kidney disease in Japanese individuals by a genome-wide
association study. J Med Genet. 50:410–418. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada Y, Matsuo H, Segawa T, et al:
Assessment of genetic risk for myocardial infarction. Thromb
Haemost. 96:220–227. 2006.PubMed/NCBI
|
|
22
|
Fujimaki T, Kato K, Yoshida T, et al:
Association of genetic variants with myocardial infarction in
Japanese individuals with chronic kidney disease. Thromb Haemost.
101:963–968. 2009.PubMed/NCBI
|
|
23
|
Oguri M, Kato K, Yokoi K, et al:
Association of genetic variants with myocardial infarction in
Japanese individuals with metabolic syndrome. Atherosclerosis.
206:486–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Go AS, Mozaffarian D, Roger VL, et al:
Heart disease and stroke statistics - 2014 update: A report from
the American Heart Association. Circulation. 129:e28–e292. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sacco RL, Benjamin EJ, Broderick JP, et
al: American Heart Association Prevention Conference. IV.
Prevention and Rehabilitation of Stroke. Risk factors. Stroke.
28:1507–1517. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamagata K, Ishida K, Sairenchi T,
Takahashi H, Ohba S, Shiigai T, Narita M and Koyama A: Risk factors
for chronic kidney disease in a community-based population: A
10-year follow-up study. Kidney Int. 71:159–166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamada Y, Matsui K, Takeuchi I, Oguri M
and Fujimaki T: Association of genetic variants with hypertension
in a longitudinal population-based genetic epidemiological study.
Int J Mol Med. (In press).
|
|
28
|
Shimokata S, Oguri M, Fujimaki T, Horibe
H, Kato K and Yamada Y: Association between polymorphisms of the
α-kinase 1 gene and type 2 diabetes mellitus in community-dwelling
individuals. Biomed Rep. 1:940–944. 2013.PubMed/NCBI
|
|
29
|
Ueyama C, Horibe H, Fujimaki T, Oguri M,
Kato K and Yamada Y: Association of genetic variants of CELSR1 and
3q28 with hypertension in community-dwelling individuals. Biomed
Rep. 1:840–844. 2013.PubMed/NCBI
|
|
30
|
Oguri M, Fujimaki T, Horibe H, Kato K,
Ichihara S and Yamada Y: Association of a polymorphism of BTN2A1
with chronic kidney disease in community-dwelling individuals.
Biomed Rep. 1:868–872. 2013.PubMed/NCBI
|
|
31
|
Fujimaki T, Horibe H, Oguri M, Kato K and
Yamada Y: Association of genetic variants of the α-kinase 1 gene
with myocardial infarction in community-dwelling individuals.
Biomed Rep. 2:127–131. 2014.PubMed/NCBI
|
|
32
|
Horibe H, Ueyama C, Fujimaki T, Oguri M,
Kato K, Ichihara S and Yamada Y: Association of a polymorphism of
BTN2A1 with dyslipidemia in community-dwelling individuals. Mol Med
Rep. 9:808–812. 2014.PubMed/NCBI
|
|
33
|
Murakata Y, Fujimaki T and Yamada Y:
Association of a butyrophilin, subfamily 2, member A1 gene
polymorphism with hypertension. Biomed Rep. 2:818–822.
2014.PubMed/NCBI
|
|
34
|
Kuzuya T, Nakagawa S, Satoh J, et al:
Report of the Committee on the classification and diagnostic
criteria of diabetes mellitus. Diabetes Res Clin Pract. 55:65–85.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Itoh Y, Mizuki N, Shimada T, Azuma F,
Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M and Inoko H:
High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a
PCR-SSOP-Luminex method in the Japanese population. Immunogenetics.
57:717–729. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hanley JA, Negassa A, Edwardes MD and
Forrester JE: Statistical analysis of correlated data using
generalized estimating equations: An orientation. Am J Epidemiol.
157:364–375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dean CB and Nielsen JD: Generalized linear
mixed models: A review and some extensions. Lifetime Data Anal.
13:497–512. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heine M, Cramm-Behrens CI, Ansari A, Chu
HP, Ryazanov AG, Naim HY and Jacob R: Alpha-kinase 1, a new
component in apical protein transport. J Biol Chem.
280:25637–25643. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang SJ, Tu HP, Ko AM, Chiang SL, Chiou
SJ, Lee SS, Tsai YS, Lee CP and Ko YC: Lymphocyte α-kinase is a
gout-susceptible gene involved in monosodium urate
monohydrate-induced inflammatory responses. J Mol Med (Berl).
89:1241–1251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kahn SE: The relative contributions of
insulin resistance and beta-cell dysfunction to the pathophysiology
of Type 2 diabetes. Diabetologia. 46:3–19. 2003.PubMed/NCBI
|
|
41
|
Fukushima M, Usami M, Ikeda M, Nakai Y,
Taniguchi A, Matsuura T, Suzuki H, Kurose T, Yamada Y and Seino Y:
Insulin secretion and insulin sensitivity at different stages of
glucose tolerance: A cross-sectional study of Japanese type 2
diabetes. Metabolism. 53:831–835. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ruotsalainen E, Salmenniemi U, Vauhkonen
I, Pihlajamӓki J, Punnonen K, Kainulainen S and Laakso M: Changes
in inflammatory cytokines are related to impaired glucose tolerance
in offspring of type 2 diabetic subjects. Diabetes Care.
29:2714–2720. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Olefsky JM and Glass CK: Macrophages,
inflammation, and insulin resistance. Annu Rev Physiol. 72:219–246.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Norlin S, Ahlgren U and Edlund H: Nuclear
factor-κB activity in β-cells is required for glucose-stimulated
insulin secretion. Diabetes. 54:125–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shriner D, Adeyemo A and Rotimi CN: Joint
ancestry and association testing in admixed individuals. PLoS
Comput Biol. 7:e10023252011. View Article : Google Scholar : PubMed/NCBI
|