Introduction
Chronic hepatitis B (CHB) imposes a major health and
economic burden as to 2 billion people worldwide have a history of
hepatitis B virus (HBV) infection and ~360 million suffering from
chronic HBV infection despite its declining incidence, leading the
main cause of chronic diseases-related malfunction (1,2). CHB may
increase the risk of developing liver cirrhosis, severe liver
failure and hepatocellular carcinoma, although primary HBV
infections usually have a self-limited course in adults (3). Due to the residual virus and weakened
immunity to reinfection, ~20% do not recover but progress to liver
cirrhosis and 5% develop hepatocellular carcinoma through
persistent infections (3,4). However, the precise mechanisms leading to
the chronicity of HBV infection remain to be elucidated at the
molecular level. Infection may spread in a variety of ways
including vertical (mother-to-child transmission) and horizontal
transmission (lesions, bites, sanitary habits, sexual contact,
medical exposure and drug use). In Asian countries, over half of
the CHB patients were infected via vertical transmission and
subsequently became HBV carriers (5).
In adolescence, ~5% of the primary HBV carriers exhibit a long-term
liver dysfunction and progress to chronic hepatitis (6). CHB significantly increases the
probability of liver cirrhosis and primary hepatocellular cancer in
the decades following the initial diagnosis and treatment (7,8).
Currently, CHB remains a major concern regarding the
issue of public health. However, the detailed pathogenesis of such
a disease remains to be elucidated. In addition to the differences
in the viral and environmental factors, the variations of host
genetic factors are proved to dominate the pathological states of
CHB development and progression. A number of genetic studies
provide evidence that variations at the genetic level contribute to
the development of chronic hepatitis (9–11). In
addition to the aforementioned evidence, extensive epidemiological
studies have shown that the variations of genetic factors,
including cytokines (12–14), human leukocyte antigen (HLA) (15–18) and
immune response-associated genes (19–21), could
evidently affect the clinical outcomes of primary HBV infection.
The HLA complex is the first discovered genetic factor exhibiting a
definite correlation with HBV infection. HLA polymorphisms are
usually associated with immune response variability. The genotype
of the HLA genes may affect the progression or regression of HBV
infection. The main function of HLA-II molecules is to present
specific antigens to cluster of differentiation 4+
(CD4+) T cells, which regulate the immune response of
CD8+ cytotoxic T lymphocytes (CTL) and are important to
the production of specific-neutralizing antibodies. The process of
HBV clearance is governed by eliminating infected cells via CTL and
protecting additional cells from persistent infection via
neutralizing antibody.
Therefore, it appears biologically viable to assume
that variability in the interaction between HLA-II molecules and
HBV antigens may be extremely important. This is verified by the
evidence that patients with acute HBV infections showed superior
HLA-II restricted CD4+ T-cell immune responses to the
hepatitis B core antigen compared with chronic hepatitis patients
(22). HLA class II gene polymorphisms
are associated with various diseases, particularly for autoimmune
disorders (23). However, the
association of the HLA class II gene polymorphism with human
diseases exhibits ethnical and geographic variability (24). Acute hepatitis B patients with strong
HLA class I and II-restricted T-cell responses will not suffer from
persistent HBV infection, while those without these responses may
progress to CHB (25–27). Shi et al (28) indicated that HLA-II genes polymorphisms
may be a crucial factor in affecting the outcome of HBV
infection.
Kamatani et al (29) demonstrated that variants in the
HLA-DP locus were strongly associated with CHB in the Asian
population by conducting a genome-wide association study (GWAS).
HLA-DQB1 polymorphisms have recently been proved to affect
immune responses of patients, and thus influence the clinical
outcome of numerous diseases (30,31). A
previous GWAS study conducted by Mbarek et al (32) suggested that there was a strong
association between the HLA-DQB1 polymorphism (rs2856718)
and CHB. HLA-DQB1*0301 is also correlated with
susceptibility to CHB (33–35), whereas HLA-DQB1*0201 is proved
to be a HBV-resistance gene in Xinjiang Uygur (35), and HLA-DQB1*0501 has been
revealed to be associated with persistent response to interferon
treatment in chronic hepatitis C patients (36). Li et al (37) suggested that HLA-DQB1*0302 could
reduce the incidence of hepatocellular carcinoma by inhibiting the
replication of HBV.
However, according to previous studies, it remains
unclear whether HLA-DQB1 polymorphisms are associated with
the susceptibility to CHB due to small sample size and small
phenotypic effects of HLA-DQB1 locus. Therefore, the present
study conducted a comprehensive meta-analysis to evaluate the
potential association between HLA-DQB1 polymorphisms and
susceptibility to CHB. HLA-DQB1 polymorphisms were
quantitatively summarized in serum samples from patients with
chronic hepatitis B infection. The case-control studies were
adopted to evaluate whether HLA-DQB1 polymorphisms are
associated with the risk of chronic HBV infection by a comparison
of the frequency distribution differences in 13 HLA-DQB1
locus between the CHB and healthy control groups.
Materials and methods
Search strategy and selection
criteria
The PubMed, Embase, CNKI and Wanfang databases were
searched for studies that reported on the association of
HLA-DQB1 polymorphisms with CHB between January 1, 1966 and
July 30, 2015, using Medical Subject Heading terms ‘major
histocompatibility complex, class II, DQβ1’ and ‘polymorphisms’ and
‘chronic hepatitis B’ or ‘chronic hepatitis B infection’ or
‘chronic hepatitis’ and corresponding free words. The Cochrane
library (http://www.cochrane.org) was also
searched using the term ‘major histocompatibility complex, class
II, DQβ1’, ‘polymorphisms’ and ‘chronic hepatitis B’ or ‘chronic
hepatitis B infection’. Furthermore, the citations of the retrieved
studies were reviewed in order to search for additional studies in
association with the present meta-analysis. Included studies met
the following criteria: i) Case-control studies, nested
case-control studies or cohort studies; ii) studies investigating
the correlation between HLA-DQB1 polymorphisms and CHB, and
the exposed risk factor should be HLA-DQB1 polymorphisms;
iii) relevant genotype frequencies, or odds ratio (OR) and 95%
confidence interval (CI) should be reported; iv) full-text studies
so that detailed information could be acquired. Excluded studies
were: i) Studies without healthy control subjects; ii) duplicated
publications; iii) studies that involved <20 participants. When
there was more than one study on the same subjects, only the most
recent study was used.
Data extraction
Data extraction was independently performed by two
experienced investigators (J. Huang and Z. Zhou) and examined
carefully by the other investigators. The concordance rate of the
investigators was 95.6%. Disagreement was resolved by consensus.
The following data was extracted from the included studies: The
first author's name, date of publication, region, ethnicity, design
method, genotyping, case and control subjects number, and genotype
frequencies. Data were collected only for subjects whose
HLA-DQB1 polymorphisms status had been detected in CHB and
its control.
Assessment of study quality
Two investigators (J. Huang and Z. Zhou)
independently assessed the quality of each included study according
to a 12-point scoring system (38).
Study design, number of cases, source of subjects, genotyping
method and matching method of case and control were examined in the
assessment of study quality. Studies, which met each of the
following criteria (a prospective study, >100 cases, including
community-based participants, DNA sequencing was used to detect
HLA-DQB1 polymorphisms, and matched for age and gender),
were scored on a 2-point scale. Studies with a total score of ≥8
were defined as high-quality studies, 5–7 were defined as
medium-quality studies, and ≤4 were regard as low-quality studies.
These cut-off values were confirmed based on the quality scores
distribution of all studies. The Spearmans rank correlation
coefficient of consensus between each of the two reviewers on the
total quality assessment for all the associated studies was 0.97.
In addition, disagreements were settled by consultation.
Statistical analysis
The meta-analysis was performed using the software
Stata 12.0 (Stata Corporation, College Station, TX, USA). OR and
its corresponding 95% CI were adopted as the effect measures to
conduct the meta-analysis. The Q-test, P-value and I2
test was used to evaluate heterogeneity among studies (39,40). When
the P-value of heterogeneity value was >0.05 and
I2<50%, a fixed-effects model was adopted to
calculate OR and its 95% CI, otherwise a random-effects model was
used. The combined OR was calculated by two-sided Z-test, and
P<0.05 was considered to indicate a statistically significance
difference. Sensitivity analysis was performed to assess the
reliability and stability of the overall results. Publication bias
test was performed using Begg's funnel plots and the Egger's
regression plots (41,42).
Results
Characteristics of eligible
studies
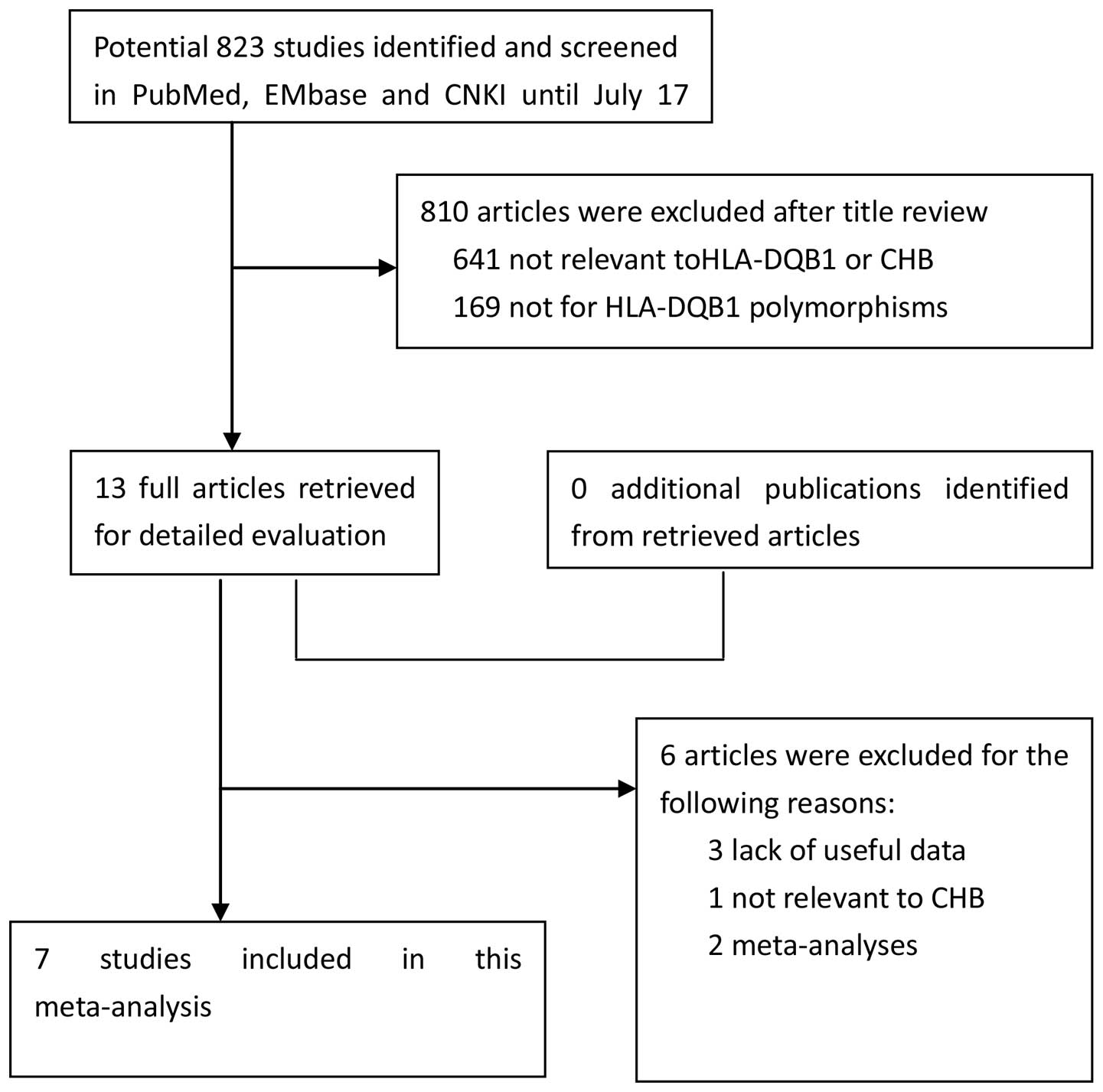
Subsequent to searching the previously defined
databases, a total of 13 studies were selected according to the
established search strategy. Six studies that were not eligible, as
shown by the data provided in the abstract and text, were excluded.
Finally, a total of 7 case-control studies were available in this
meta-analysis, including 815 CHB patients and 731 control subjects
for HLA-DQB1 polymorphisms (34,35,37,43–46). The study search and selection process
is shown in Fig. 1. The detailed
characteristics of the 7 included studies are shown in Table I. The publication year of the included
studies ranged between 2003 and 2015. The distribution of the
HLA-DQB1 polymorphisms in CHB is shown in Table II. All the studies used blood samples
for HLA-DQB1 genotyping. All the quality scores of the
included studies were >7 (moderate-high quality) (38).
 | Table I.Characteristics of the studies
included in the meta-analysis. |
Table I.
Characteristics of the studies
included in the meta-analysis.
| First author
(year) | Region | Ethnicity | Design | Genotyping | Case (n) | Control (n) | Refs. |
|---|
| Jiang et al
(2003) | China | Asian | PB | PCR/SSP | 52 | 106 | (34) |
| Park et al
(2003) | Korean | Asian | PB | PCR/RFLP/SSCP | 135 | 100 | (43) |
| Xi-Lin et al
(2006) | China | Asian | HB | PCR/SSP | 139 | 134 | (44) |
| Liu and Cheng
(2007) | China | Asian | PB | PCR/SSP | 168 | 100 | (45) |
| Zhu et al
(2007) | China | Asian | HB | PCR/SSP | 151 | 133 | (46) |
| Zhang et al
(2015) | China | Asian | HB | PCR/SSP | 110 | 100 | (35) |
| Li et al
(2015) | China | Asian | HB | PCR/SSP | 60 | 58 | (37) |
 | Table II.Distribution of the HLA-DQB1
polymorphisms in chronic hepatitis B. |
Table II.
Distribution of the HLA-DQB1
polymorphisms in chronic hepatitis B.
|
|
HLA-DQB1 loci |
|
|---|
|
|
|
|
|---|
| First author
(year) | 0201 | 0301 | 0302 | 0303 | 0401 | 0402 | 0501 | 0502 | 0503 | 0601 | 0602 | 0603 | 0604 | Refs. |
|---|
| Jiang et al
(2003) |
|
|
|
|
|
|
|
|
|
|
|
|
| (34) |
| Case,
n | 10 | 37 | 6 | 15 | 5 | 1 | 3 | 7 | 2 | 7 | 4 | 2 | 2 |
|
|
Control, n | 23 | 40 | 14 | 35 | 11 | 2 | 9 | 20 | 6 | 20 | 12 | 5 | 7 |
|
| Park et al
(2003) |
|
|
|
|
|
|
|
|
|
|
|
|
| (43) |
| Case,
n | 31 | 43 | 27 | 23 | 22 | 5 | 12 | 9 | 17 | 31 | 27 | 2 | 4 |
|
|
Control, n | 13 | 26 | 18 | 24 | 9 | 11 | 17 | 3 | 11 | 12 | 18 | 5 | 14 |
|
| Xi-Lin et al
(2006) |
|
|
|
|
|
|
|
|
|
|
|
|
| (44) |
| Case,
n | 48 | 71 | 10 | 58 | 8 | NA | 12 | 17 | 8 | 24 | 21 | 1 | 0 |
|
|
Control, n | 45 | 55 | 9 | 74 | 10 | NA | 9 | 11 | 14 | 25 | 13 | 0 | 3 |
|
| Liu and Cheng
(2007) |
|
|
|
|
|
|
|
|
|
|
|
|
| (45) |
| Case,
n | 63 | 10 | 14 | 14 | 9 | 5 | 7 | 9 | 16 | 45 | 67 | 7 | 7 |
|
|
Control, n | 17 | 5 | 8 | 12 | 10 | 5 | 2 | 3 | 8 | 15 | 52 | 3 | 7 |
|
| Zhu et al
(2007) |
|
|
|
|
|
|
|
|
|
|
|
|
| (46) |
| Case,
n | 51 | 79 | 12 | 61 | 8 | NA | 15 | 18 | NA | 26 | 22 | NA | NA |
|
|
Control, n | 38 | 56 | 20 | 61 | 10 | NA | 18 | 3 | NA | 15 | 25 | NA | NA |
|
| Zhang et al
(2015) |
|
|
|
|
|
|
|
|
|
|
|
|
| (35) |
| Case,
n | 20 | 30 | 9 | 22 | 3 | NA | 5 | 7 | NA | NA | 10 | 1 | 1 |
|
|
Control, n | 11 | 17 | 8 | 31 | 4 | NA | 6 | 5 | NA | NA | 9 | 2 | 1 |
|
| Li et al
(2015) |
|
|
|
|
|
|
|
|
|
|
|
|
| (37) |
| Case,
n | 3 | 42 | 2 | 13 | 6 | 1 | 6 | 7 | 4 | 11 | 8 | 1 | NA |
|
|
Control, n | 8 | 27 | 8 | 17 | 3 | 0 | 6 | 8 | 3 | 16 | 7 | 1 | NA |
|
Quantitative data synthesis
In conslusion, statistically significant pooled OR
of HLA-DQB1 polymorphisms were obtained for HLA-DQB1
loci [*0201, case vs. control: I2=36.5%; P-value of
heterogeneity=0.15; OR, 1.29; 95% CI, 1.02–1.64; P=0.0301 (Fig. 2A and Table
III); *0301, case vs. control: I2=0%; P-value of
heterogeneity=0.899; OR, 1.37; 95% CI, 1.12–1.69; P=0.002 (Fig. 2B and Table
III); *0502, case vs. control: I2=24.9%; P-value of
heterogeneity=0.239; OR, 1.50; 95% CI, 1.02–2.20; P=0.04 (Fig. 2C and Table
III)], which were associated with increased risk of CHB.
Similar significant results were observed and acquired in the
following HLA-DQB1 loci [*0303, case vs. control:
I2=0%; P-value of heterogeneity=0.986; OR, 0.77; 95% CI,
0.62–0.95; P=0.017 (Fig. 2D and
Table III); *0604, case vs. control:
I2=0%; P-value of heterogeneity=0.594; OR, 0.38; 95% CI,
0.20–0.74; P=0.003 (Fig. 2E and
Table III)], which were associated
with a decreased risk of CHB. No significant association was
observed for the other HLA-DQB1 family loci (Table III).
 | Table III.Results of Q-test and I2
test for HLA-DQB1 polymorphisms in chronic hepatitis B. |
Table III.
Results of Q-test and I2
test for HLA-DQB1 polymorphisms in chronic hepatitis B.
| HLA-DQB1
loci | Q-value | I2,
% | P-value | OR (95% CI) | Pooled P-value |
|---|
| 0201 |
9.45 | 36.5 | 0.150 | 1.29
(1.02–1.64) | 0.031 |
| 0301 |
2.21 |
0.0 | 0.899 | 1.37
(1.12–1.69) | 0.002 |
| 0302 |
5.12 |
0.0 | 0.523 | 0.84
(0.60–1.16) | 0.290 |
| 0303 |
0.98 |
0.0 | 0.986 | 0.77
(0.62–0.95) | 0.017 |
| 0401 | 53.23 | 88.7 | <0.001 | 0.47
(0.15–1.43) | 0.182 |
| 0402 |
2.05 |
0.0 | 0.562 | 0.53
(0.25–1.10) | 0.088 |
| 0501 |
3.81 |
0.0 | 0.702 | 0.82
(0.57–1.19) | 0.293 |
| 0502 |
7.98 | 24.9 | 0.239 | 1.50
(1.02–2.20) | 0.040 |
| 0503 |
2.17 |
0.0 | 0.704 | 0.93
(0.60–1.45) | 0.741 |
| 0601 |
7.39 | 32.3 | 0.193 | 1.24
(0.93–1.64) | 0.138 |
| 0602 |
3.67 |
0.0 | 0.721 | 0.93
(0.72–1.20) | 0.565 |
| 0603 |
2.84 |
0.0 | 0.725 | 0.79
(0.38–1.66) | 0.536 |
| 0604 |
2.78 |
0.0 | 0.594 | 0.38
(0.20–0.74) | 0.003 |
Sensitivity analysis
Sensitivity analysis was performed by removing one
study at a time to detect the source of heterogeneity. There was no
evident heterogeneity in all the HLA-DQB1 family loci.
Additionally, there was no valid evidence to support that any study
independently influenced the combined OR, which indicated that the
overall results of this study are robust and convincing, as shown
in the plots for sensitivity analysis (Fig. 3).
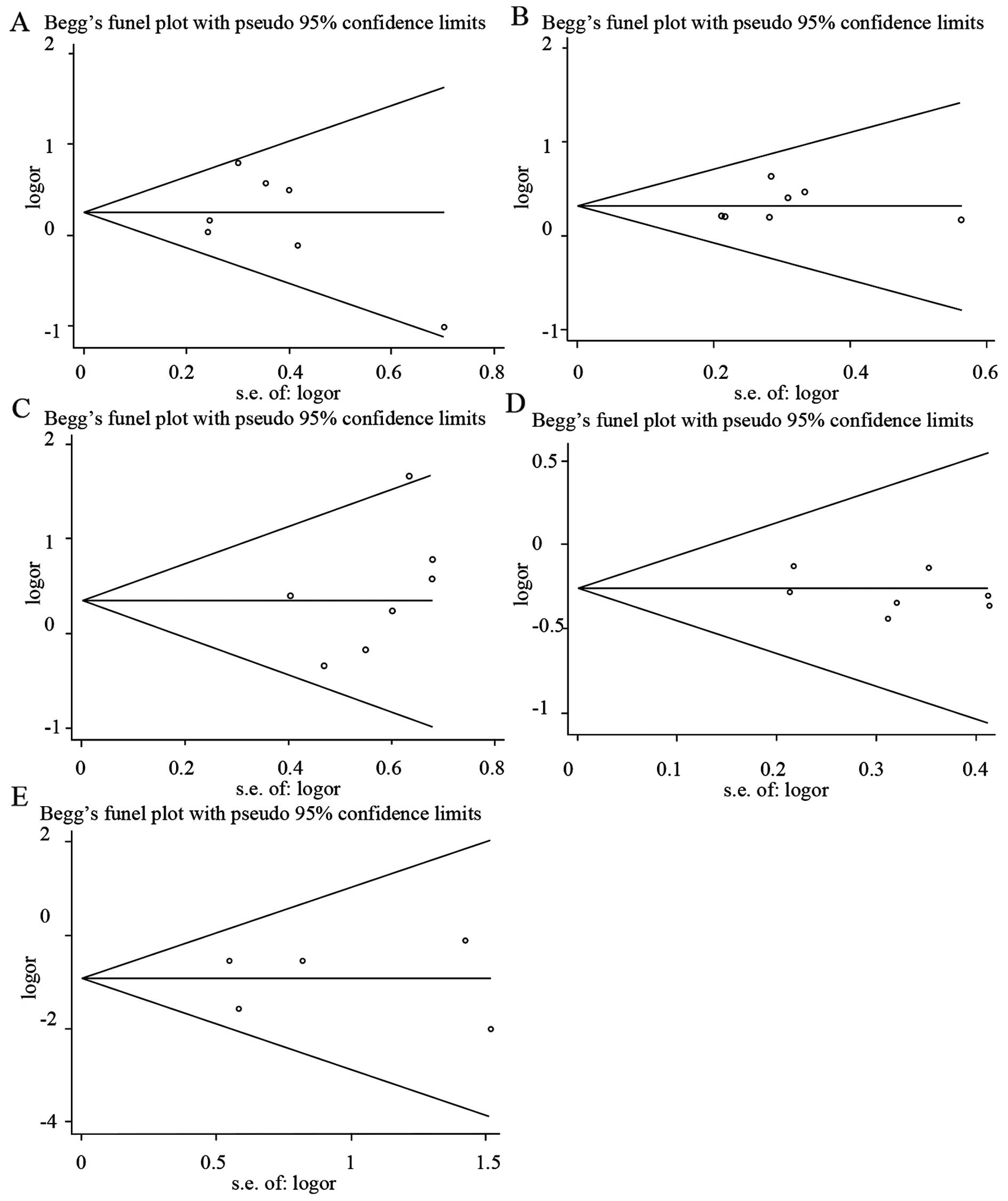
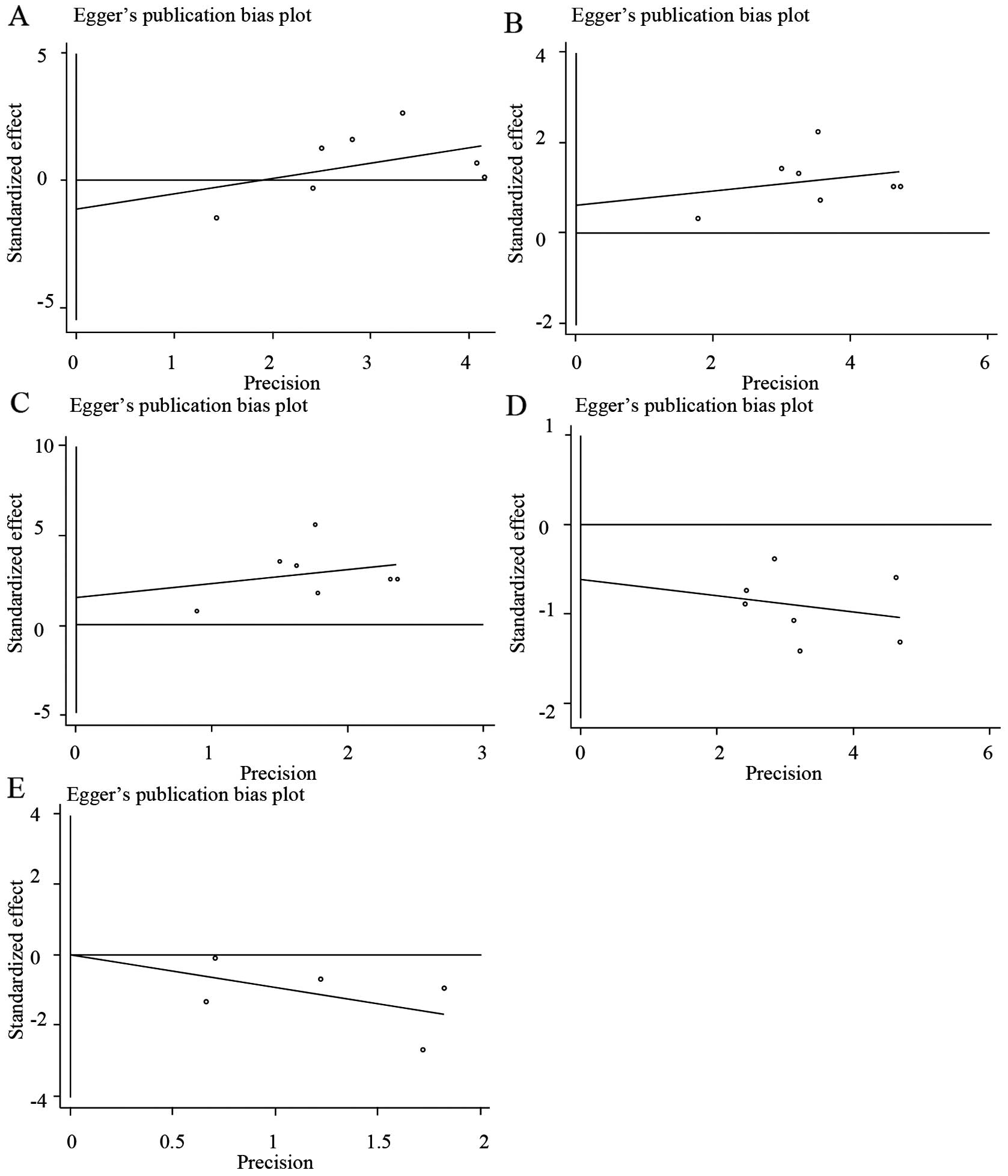
Publication bias
Begg's funnel plots and Egger's regression plots
were used to detect the publication bias of all the HLA-DQB1
loci. As illustrated in Fig. 4, the
funnel plots did not show any evidence of significant asymmetry and
suggested that no publication bias existed [*0201, Z=0.6, P=0.548
(Fig. 4A and Table IV); *0301, Z=1.5, P=0.133 (Fig. 4B and Table
IV); *0502, Z=1.5, P=0.133 (Fig.
4C and Table IV); *0303, Z=0, P=1
(Fig. 4D and Table IV); *0604, Z=0.24, P=0.86 (Fig. 4E and Table
IV)]. Egger's test also indicated that there was no
statistically significant publication bias [*0201, T=−0.66, P=0.54
(Fig. 5A and Table IV); *0301, T=0.63, P=0.554 (Fig. 5B and Table
IV); *0502, T=1.18, P=0.291 (Fig.
5C and Table IV); *0303, T=−1.03,
P=0.349 (Fig. 5D and Table IV); *0604, T=0.01, P=0.996 (Fig. 5E and Table
IV)].
 | Table IV.Results of Beggs test and Eggers test
for HLA-DQB1 polymorphisms in chronic hepatitis B. |
Table IV.
Results of Beggs test and Eggers test
for HLA-DQB1 polymorphisms in chronic hepatitis B.
|
| Beggs test | Eggers test |
|---|
|
|
|
|
|---|
| HLA-DQB1 loci | Z-value | P-value | T-value | Pooled P-value |
|---|
| 0201 | 0.60 | 0.548 | −0.66 | 0.540 |
| 0301 | 1.50 | 0.133 | 0.63 | 0.554 |
| 0302 | 1.50 | 0.133 | −1.24 | 0.270 |
| 0303 | <0.01 | 1.000 | −1.03 | 0.349 |
| 0401 | <0.01 | 1.000 | −0.25 | 0.811 |
| 0402 | 1.70 | 0.089 | 3.57 | 0.070 |
| 0501 | 0.90 | 0.368 | 1.30 | 0.249 |
| 0502 | 1.50 | 0.133 | 1.18 | 0.291 |
| 0503 | 0.73 | 0.462 | −0.14 | 0.897 |
| 0601 | 0.38 | 0.707 | −1.24 | 0.282 |
| 0602 | 0.60 | 0.548 | 0.80 | 0.459 |
| 0603 | 0.38 | 0.707 | 0.22 | 0.836 |
| 0604 | 0.24 | 0.806 | 0.01 | 0.996 |
Discussion
To the best of our knowledge, this is the first
study investigating the association of HLA-DQB1 alleles with
CHB. Numerous studies have suggested the associations of HLA
gene polymorphisms with inflammatory diseases and autoimmune
diseases, such as HBV infection (32,47),
hepatitis C virus infection (48,49),
systemic lupus erythematosus (50) and
rheumatoid arthritis (51). However,
the majority of these studies focus on the correlation between the
HLA antigen and CHB based on a small sample size and HLA serotyping
that has limited resolution; therefore, those results may be
inaccurate and inconsistent for the distribution of numerous
HLA-DQB1 loci. Along with the development of genotyping
methods, HLA-genotyping is becoming more precise in the
identification of the HLA-DQB1 loci, and more accurate in
the identification of the peptide-binding site of MHC II molecules.
Therefore, HLA genotyping methods are being used more frequently in
the study of immunogenetics.
Recent studies on the correlation between
HLA-DQB1 polymorphisms and CHB have been inconsistent and
inconclusive. Jiang et al (34)
reported that HLA-DQB1*0301 are closely associated with
susceptibility to CHB, while other HLA-DQB1 alleles are not.
Thus, it is plausible that the HLA-DQB1*0301 allele may be a
risk factor for the development of CHB (OR, 3.9). Park et al
(43) insisted that
HLA-DQB1*0402 and DQB1*0604 alleles have a certain
protective effect on the occurrence of CHB (OR, 0.3; and OR, 0.1,
respectively). Therefore, these alleles may be considered as good
prognostic factors. Liu and Cheng (45) observed that the HLA-DQB1*0201
and DQB1*0601 alleles have significant susceptible effect on
chronic HBV infection (OR, 2.93; and OR, 2.07, respectively).
However, Xi-Lin et al (44)
identified that the HLA-DQB1*0303 and DQB1*0503
alleles are independently resistant genetic factors to CHB (OR,
0.65; and OR, 0.35, respectively). Zhu et al (46) further observed that the
HLA-DQB1*0502 allele is significantly associated with the
clinical outcome of HBV infection (OR, 18) and is a host genetic
risk factor for HBV infection.
The present study showed that five specific
HLA-DQB1 loci are associated with an increased or decreased
risk of CHB. Among the 13 specific HLA-DQB1 alleles,
DQB1*0201, DQB1*0301 and DQB1*0502 were
significantly associated with the increased risk of CHB. The pooled
OR was 1.29 (95% CI, 1.02–1.64; P=0.0301), 1.37 (95% CI, 1.12–1.69;
P=0.002) and 1.50 (95% CI, 1.02–2.20; P=0.04), respectively.
However, DQB1*0303 and DQB1*0604 were significantly
associated with a decreased risk of CHB. The pooled OR was 0.77
(95% CI, 0.62–0.95; P=0.017) and 0.38 (95% CI, 0.20–0.74; P=0.003),
respectively. No significant association was observed for the other
HLA-DQB1 family alleles. The overall results indicate that
HLA-DQB1*0201, HLA-DQB1*0301 and HLA-DQB1*0502
alleles may have a significantly higher risk for CHB, while
HLA-DQB1*0303 and HLA-DQB1*0604 may have a
significantly protective effect for CHB.
The study by Zhang et al (35) suggested that HLA-DQB1*0303 is a
resistance gene of CHB in Xinjiang Uygur, while
HLA-DQB1*0301 is associated with continuous infection of
HBV. The HLA-DQB1*0201 distribution frequency in the low
copy group was significantly higher than that of the high copy
group (OR, 1.939; P<0.05), and thus assumed that
DQB1*0201 may contribute to the clearance of HBV (35). In addition, Li et al (52) reported that the HLA-DQB1*0501,
HLA-DQB1*0601 and HLA-DQB1*0602 alleles are
associated with significantly increased immunological responses to
the hepatitis B vaccine in healthy people (OR, 1.85; OR, 2.35; and
OR, 2.34, respectively), while HLA-DQB1*0201 is adverse (OR,
0.27). The mechanisms underlying these effects on CHB are not fully
elucidated, but larger-scale studies provide a promise of further
confirmation. Jiang et al (53)
identified five novel susceptibility loci for CHB using a GWAS with
2,514 CHB cases and 1,130 normal controls from eastern China, and
four of them are located in the human leukocyte antigen (HLA)
region at 6p21.3. Additionally, the study validated seven
previously reported CHB susceptibility loci, including rs2856718 at
HLA-DQB1, rs7453920 at HLA-DQB2, rs3077 at HLA-DPA1,
rs9277535 at HLA-DPA2, rs3130542 at HLA-C, rs1419881 at TCF19, and
rs652888 at EHMT2 (53). All are
located in the HLA region.
CHB development is preceded by acute inflammation
and immune responses. Whether antigen-presenting cells are able to
identify HBV antigens may be critical for the development of CHB.
The correlation of specific HLA-DQB1 alleles with resistance
or susceptibility to CHB is possibly attributed to a direct effect
of HLA-DQB1 molecule as an antigen-presenting unit or
possibly owing to a neighboring-related gene (53). We assume that the host immune response
status of the patients with CHB and carrying HLA-DQB1
polymorphisms are changed. T-cells are often activated under
certain conditions such as infection, depression and fatigue.
Accompanied by the removal of HBV, liver damage was triggered and a
range of clinical symptoms occurred such as fever, anorexia,
abnormally elevated aminotransferases and icterus, inducing the
formation of CHB. With regards to the HLA-DQB1 loci, it may
be plausible that HLA molecule mediates the function of host
antigen-presenting cells and induces cytotoxic T-lymphocyte
responses.
However, due to the potential heterogeneity of HBV,
the results of the present study should be explained with caution.
These retrospective studies are more prone to bias than prospective
randomized clinical trial (RCT) studies. The information of CHB
patients complicated by HCC were not specially extracted and
analyzed. The association of HLA-DQB1 loci with HCV
infection was not included in the meta-analysis. The overall sample
size was relatively small due to the limited number of original
studies.
In conclusion, the present meta-analysis suggests
that HLA-DQB1*0201, DQB1*0301 and DQB1*0502
are risk factors for CHB, while HLA-DQB1*0303 and
DQB1*0604 are protective factors. These results are
compatible with the published studies regarding the correlation
between HLA-DQB1 loci and other inflammatory disorders.
Future large scale studies of HLA-DQB1 should be used to
provide strong evidence for a genetic contribution to CHB.
References
|
1
|
Ocama P, Opio CK and Lee WM: Hepatitis B
virus infection: Current status. Am J Med. 118:14132005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ott JJ, Stevens GA, Groeger J and Wiersma
ST: Global epidemiology of hepatitis B virus infection: New
estimates of age-specific HBsAg seroprevalence and endemicity.
Vaccine. 30:2212–2219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ganem D and Prince AM: Hepatitis B virus
infection - natural history and clinical consequences. N Engl J
Med. 350:1118–1129. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wright TL and Lau JY: Clinical aspects of
hepatitis B virus infection. Lancet. 342:1340–1344. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pungpapong S, Kim WR and Poterucha JJ:
Natural history of hepatitis B virus infection: An update for
clinicians. Mayo Clin Proc. 82:967–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okada K, Kamiyama I, Inomata M, Imai M,
Miyakawa Y and Mayumi M: e antigen and anti-e in the serum of
asymptomatic carrier mothers as indicators of positive and negative
transmission of hepatitis B virus to their infants. N Engl J Med.
294:746–749. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chemin I and Zoulim F: Hepatitis B virus
induced hepatocellular carcinoma. Cancer Lett. 286:52–59. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McMahon BJ: The natural history of chronic
hepatitis B virus infection. Hepatology. 49:S45–S55. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS,
Lin CC, Kwang TY, Hsu ST, Lin SY and Hsu LC: Hepatitis B virus
markers in Chinese twins. Anticancer Res. 9:737–741.
1989.PubMed/NCBI
|
|
10
|
Tong H, Bock CT and Velavan TP: Genetic
insights on host and hepatitis B virus in liver diseases. Mutat Res
Rev Mutat Res. 762:65–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su M, Zeng Y, Chen J, Jiang L, Chen T, Liu
C, Yang B and Ou Q: Studies on the association of single nucleotide
polymorphisms of HLA-DP and DQ genes with the outcome of chronic
hepatitis B virus infection. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
31:765–769. 2014.(In Chinese). PubMed/NCBI
|
|
12
|
Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes
J, Tambur AR, Tur-Kaspa R and Klein T: Cytokine gene polymorphisms
in patients infected with hepatitis B virus. Am J Gastroenterol.
98:144–150. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Höhler T, Kruger A, Gerken G, Schneider
PM, Meyer Zum Büschenefelde KH and Rittner C: A tumor necrosis
factor-alpha (TNF-alpha) promoter polymorphism is associated with
chronic hepatitis B infection. Clin Exp Immunol. 111:579–582. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Migita K, Maeda Y, Abiru S, Nakamura M,
Komori A, Miyazoe S, Nakao K, Yatsuhashi H, Eguchi K and Ishibashi
H: Polymorphisms of interleukin-1beta in Japanese patients with
hepatitis B virus infection. J Hepatol. 46:381–386. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thio CL, Carrington M, Marti D, O'Brien
SJ, Vlahov D, Nelson KE, Astemborski J and Thomas DL: Class II HLA
alleles and hepatitis B virus persistence in African Americans. J
Infect Dis. 179:1004–1006. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kummee P, Tangkijvanich P, Poovorawan Y
and Hirankarn N: Association of HLA-DRB1*13 and TNF-alpha gene
polymorphisms with clearance of chronic hepatitis B infection and
risk of hepatocellular carcinoma in Thai population. J Viral Hepat.
14:841–848. 2007.PubMed/NCBI
|
|
17
|
Hwang SH, Sohn YH, Oh HB, Hwang CY, Lee
SH, Shin ES and Lee KJ: Human leukocyte antigen alleles and
haplotypes associated with chronicity of hepatitis B virus
infection in Koreans. Arch Pathol Lab Med. 131:117–121.
2007.PubMed/NCBI
|
|
18
|
Laaribi AB, Zidi I, Hannachi N, Ben Yahia
H, Chaouch H, Bortolotti D, Zidi N, Letaief A, Yacoub S, Boudabous
A, et al: Association of an HLA-G 14-bp Insertion/Deletion
polymorphism with high HBV replication in chronic hepatitis. J
Viral Hepat. 22:835–841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thio CL, Mosbruger TL, Kaslow RA, Karp CL,
Strathdee SA, Vlahov D, O'Brien SJ, Astemborski J and Thomas DL:
Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B
virus infection. J Virol. 78:11258–11262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou J, Lu L, Yuen MF, Lam TW, Chung CP,
Lam CL, Zhang B, Wang S, Chen Y, Wu SH, et al: Polymorphisms of
type I interferon receptor 1 promoter and their effects on chronic
hepatitis B virus infection. J Hepatol. 46:198–205. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chong WP, To YF, Ip WK, Yuen MF, Poon TP,
Wong WH, Lai CL and Lau YL: Mannose-binding lectin in chronic
hepatitis B virus infection. Hepatology. 42:1037–1045. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao T, Desombere I, Vanlandschoot P,
Sällberg M and Leroux-Roels G: Characterization of HLA
DR13-restricted CD4(+) T cell epitopes of hepatitis B core antigen
associated with self-limited, acute hepatitis B. J Gen Virol.
83:3023–3033. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Battelino T, Ursic-Bratina N, Dolzan V,
Stopar-Obreza M, Pozzilli P, Krzisnik C and Vidan-Jeras B: The
HLA-DRB, -DQB polymorphism and anti-insulin antibody response in
Slovenian patients with type 1 diabetes. Eur J Immunogenet.
30:223–227. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crespí C, Milà J, Martínez-Pomar N,
Etxagibel A, Muñoz-Saa I, Priego D, Luque A, Pons J, Picornell A,
Ramon M, et al: HLA polymorphism in a Majorcan population of Jewish
descent: Comparison with Majorca, Minorca, Ibiza (Balearic Islands)
and other Jewish communities. Tissue Antigens. 60:282–291. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen WN and Oon CJ: Mutation ‘hot spot’ in
HLA class I-restricted T cell epitope on hepatitis B surface
antigen in chronic carriers and hepatocellular carcinoma. Biochem
Biophys Res Commun. 262:757–761. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kennedy PT, Sandalova E, Jo J, Gill U,
Ushiro-Lumb I, Tan AT, Naik S, Foster GR and Bertoletti A:
Preserved T-cell function in children and young adults with
immune-tolerant chronic hepatitis B. Gastroenterology. 143:637–645.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han Y, Jiang ZY, Jiao LX, Yao C, Lin QF,
Ma N, Ju RQ, Yang F, Yu JH and Chen L: Association of human
leukocyte antigen-DRB1 alleles with chronic hepatitis B virus
infection in the Han Chinese of Northeast China. Mol Med Rep.
5:1347–1351. 2012.PubMed/NCBI
|
|
28
|
Shi C, Qian YH, Su J, Luo SS, Gu J, You H,
Cui Q, Lin YD, Dong MH and Yu RB: Genetic variation in the LMP/TAP
gene and outcomes of hepatitis B virus infection in the Chinese
population. Epidemiol Infect. 139:674–682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamatani Y, Wattanapokayakit S, Ochi H,
Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N,
Kumada H, et al: A genome-wide association study identifies
variants in the HLA-DP locus associated with chronic hepatitis B in
Asians. Nat Genet. 41:591–595. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferstl B, Zacher T, Lauer B,
Blagitko-Dorfs N, Carl A and Wassmuth R: Allele-specific
quantification of HLA-DQB1 gene expression by real-time reverse
transcriptase-polymerase chain reaction. Genes Immun. 5:405–416.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Muro M, Herrero N, Marin L, Torío A,
Minguela A, Sánchez-Bueno F, García-Alonso AM and Alvarez-López MR:
Polymorphism in the upstream regulatory region of the HLA-DQB1 gene
in liver graft recipients. Hum Biol. 73:845–854. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo
M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, et al: A
genome-wide association study of chronic hepatitis B identified
novel risk locus in a Japanese population. Hum Mol Genet.
20:3884–3892. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng XQ, Chen HG, Ma YL and Liu KZ:
Influence of HLA class II molecules on the outcome of hepatitis B
virus infection in population of Zhejiang Province in China.
Hepatobiliary Pancreat Dis Int. 2:230–233. 2003.PubMed/NCBI
|
|
34
|
Jiang YG, Wang YM, Liu TH and Liu J:
Association between HLA class II gene and susceptibility or
resistance to chronic hepatitis B. World J Gastroenterol.
9:2221–2225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Zhao F, Lan L, Qin Z and Jun L:
Correlation of HLA-DQB1 gene polymorphism of Xinjiang Uygur with
outcome of HBV infection. Int J Clin Exp Med. 8:6067–6072.
2015.PubMed/NCBI
|
|
36
|
Yu ML, Dai CY, Chen SC, Chiu CC, Lee LP,
Lin ZY, Hsieh MY, Wang LY, Chuang WL and Chang WY: Human leukocyte
antigen class I and II alleles and response to interferon-alpha
treatment, in Taiwanese patients with chronic hepatitis C virus
infection. J Infect Dis. 188:62–65. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li QJ LX, Zhang LY and Zhao FL:
Correlation between polymorphisms in the human leukocyte
antigen-DQB1 alleles and hepatitis B with primary hepatocellular
carcinoma. Chin Zhonghua Gan Zang Bing Za Zhi. 23:270–274. 2015.(In
Chinese).
|
|
38
|
Liu S, Zhang H, Gu C, Yin J, He Y, Xie J
and Cao G: Associations between hepatitis B virus mutations and the
risk of hepatocellular carcinoma: A meta-analysis. J Natl Cancer
Inst. 101:1066–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bowden J, Tierney JF, Copas AJ and Burdett
S: Quantifying, displaying and accounting for heterogeneity in the
meta-analysis of RCTs using standard and generalised Q statistics.
BMC Med Res Methodol. 11:412011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Park MH, Song EY, Ahn C, Oh KH, Yang J,
Kang SJ and Lee HS: Two subtypes of hepatitis B virus-associated
glomerulonephritis are associated with different HLA-DR2 alleles in
Koreans. Tissue Antigens. 62:505–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xi-Lin Z, Te D, Jun-Hong L, Liang-Ping L,
Xin-Hui G, Ji-Rong G, Chun-Yan G, Zhuo L, Ying L and Hui L:
Analysis of HLA-DQB1 gene polymorphisms in asymptomatic HBV
carriers and chronic hepatitis B patients in the Chinese Han
population. Int J Immunogenet. 33:249–254. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu C and Cheng B: Association of
polymorphisms of human leucocyte antigen-DQA1 and DQB1 alleles with
chronic hepatitis B virus infection, liver cirrhosis and
hepatocellular carcinoma in Chinese. Int J Immunogenet. 34:373–378.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu XL, Du T, Li JH, Lu LP, Guo XH, Gao
JR, Gou CY, Li Z, Liu Y and Li H: Association of HLA-DQB1 gene
polymorphisms with outcomes of HBV infection in Chinese Han
population. Swiss Med Wkly. 137:114–120. 2007.PubMed/NCBI
|
|
47
|
Zhang X, Jia J, Dong J, Yu F, Ma N, Li M,
Liu X, Liu W, Li T and Liu D: HLA-DQ polymorphisms with HBV
infection: Different outcomes upon infection and prognosis to
lamivudine therapy. J Viral Hepat. 21:491–498. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hong X, Yu RB, Sun NX, Wang B, Xu YC and
Wu GL: Human leukocyte antigen class II DQB1*0301, DRB1*1101
alleles and spontaneous clearance of hepatitis C virus infection: A
meta-analysis. World J Gastroenterol. 11:7302–7307. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yue M, Xu K, Wu MP, Han YP, Huang P, Peng
ZH, Wang J, Su J, Yu RB, Li J, et al: Human leukocyte antigen class
II alleles are associated with hepatitis C virus natural
susceptibility in the Chinese population. Int J Mol Sci.
16:16792–16805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Castaño-Rodríguez N, Diaz-Gallo LM,
Pineda-Tamayo R, Rojas-Villarraga A and Anaya JM: Meta-analysis of
HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with
systemic lupus erythematosus. Autoimmun Rev. 7:322–330. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Laivoranta-Nyman S, Möttönen T, Hermann R,
Tuokko J, Luukkainen R, Hakala M, Hannonen P, Korpela M,
Yli-Kerttula U, Toivanen A, et al: FIN-RACo Trial Group: HLA-DR-DQ
haplotypes and genotypes in Finnish patients with rheumatoid
arthritis. Ann Rheum Dis. 63:1406–1412. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li ZK, Nie JJ, Li J and Zhuang H: The
effect of HLA on immunological response to hepatitis B vaccine in
healthy people: A meta-analysis. Vaccine. 31:4355–4361. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jiang DK, Ma XP, Yu H, Cao G, Ding DL,
Chen H, Huang HX, Gao YZ, Wu XP, Long XD, et al: Genetic variants
in five novel loci including CFB and CD40 predispose to chronic
hepatitis B. Hepatology. 62:118–128. 2015. View Article : Google Scholar : PubMed/NCBI
|