Introduction
Paraoxonase 2 (PON2) is a member of the
PON gene family located on chromosome 7q21.3, which also
includes the homologous genes for PON1 and PON3
(1,2). In
contrast to PON1, which is mainly expressed in the liver,
PON2 is expressed in a variety of tissues. It is known that
PON1 has been shown to have antioxidant properties (3). However, the function of ubiquitously
expressed PON2 remains to be elucidated (1).
A G/C single-nucleotide polymorphism (SNP) rs12026
in exon 5 of the PON2 gene introduces the coding variant of
alanine/glycine at position 148 (PON2-148). There have been
associating variants of rs12026 within the PON2 gene with
cardiovascular disease (4–6), cerebrovascular disease (7), diabetes (8,9) and other
diseases (10). For this SNP
detection, several assays have been suggested, such as the
fluorescently labeled probes technique (11), dynamic allele-specific hybridization
technique (7), TaqMan assay (5) and polymerase chain reaction-restriction
fragment length polymorphism (PCR-RFLP) (6). Among them, PCR-RFLP is the most
frequently used technique due to its high sensitivity and
reliability. However, the current PCR-RFLP assay is facing
practical challenges and is not suitable for high throughput
screening. The present study demonstrated an improved PCR-RFLP
assay for the detection of a polymorphism of the PON2
gene.
Materials and methods
Primer design
The information regarding the PON2 gene and
polymorphism rs12026 was obtained from the National Center for
Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). The method of the
amplification-created restriction site was used to introduce a new
enzyme site to recognize the variant.
The sequences of the primer pair were designed by
primer premier 5.0 software as follows: Forward, 5′
AGGTCCCGTAGTTATGTCTTGT 3′; and reverse, 5′ TCAGATGCAACAGAGAATTGTCT
3′.
The natural occurring DNA sequences according with
positions of primers were: AGGTCCCGTAGTTATGTCTTGT
aaattaactctgttctttcaattctttagatgacacagtttatctctttgttgtaaaccaccc
agaattcaagaatacagtggaaatttttaaatttgaagaagSAGAAAATTCTCTGTTGCATCTGA.
Capital bases A, C, G and T represent the sequences according to
the positions of primers, and S represents the position for
the polymorphism G/C. The length of the base sequences was 150 base
pairs (bp). The natural occurring ‘T’ nucleotide that matches with
A (underlined) in the DNA sequences was substituted by a ‘G’ in the
reverse primer (underlined) in order to introduce a recognized site
for the BsmAI restriction endonuclease.
Genotype of DNA
Genomic DNA was extracted from the peripheral blood
leukocytes of 302 Chinese Han individuals followed by standard
procedures. The research objects were obtained from one enterprise
in Zhengzhou in 2014. The study protocol was approved by the Ethics
Committee of the University of Zhengzhou (Zhengzhou, Henan, China).
The PCR was performed in a volume of 25 µl containing 100 ng of
genomic DNA, 1X PCR Master mix (Tiangen, Beijing, China) and 5 pmol
of each primer. The DNA was denatured at 95°C for 3 min, and
temperature cycles were set at 95°C for 30 sec, 56°C for 30 sec and
72°C for 30 sec for 30 cycles, followed by a final extension at
72°C for 10 min. PCR products underwent electrophoresis on 3%
agarose gels and were stained with ethidium bromide and exposed to
ultraviolet illumination.
Enzyme digestion was conducted in a 20 µl final
volume using 5 units of BsmAI enzyme (New England Biolabs,
Ipswich, MA, USA) and 10 µl of the PCR product. The reaction was
conducted at 37°C overnight. The digested products were visualized
on 3% agarose gels stained with ethidium bromide.
Statistical analysis
The basic data were analyzed using SPSS 21.0
software (IBM, Corp., Armonk, NY, USA). Methods of representation
and examination were based on the distribution of quantitative
data. All the statistical tests were two-sided, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Polymorphisms of PON2
Based on the PCR designed with the mismatched
primers, the PON2 polymorphisms could be identified
simultaneously. The PCR products that were separated on 3% agarose
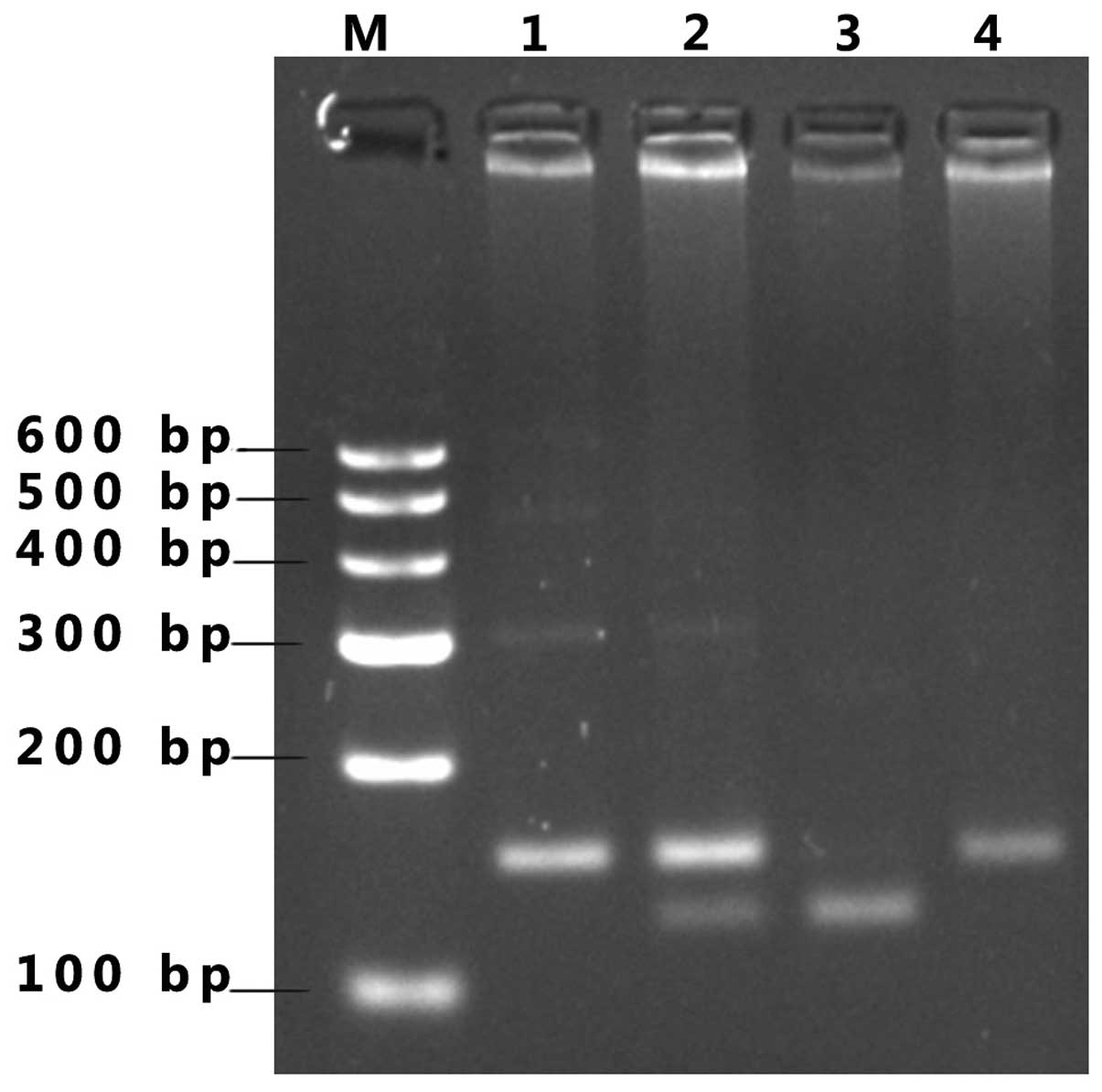
gel are shown in Fig. 1 (BsmAI
PCR-RFLP analysis of the PON2 biallelic polymorphism). From
the gel, that the PCR amplification yielded a product of 150 bp.
Following incubation with BsmAI, the individual homozygous
for the C allele yielded one uncut band (150 bp). The homozygous G
allele yielded two bands of 121 and 29 bp.
Polymorphisms of PON2 were detected in 302
Chinese Han individuals. The genotype frequencies were 68.9% for
CC, 29.8% for CG and 1.3% for GG. The allelic frequencies were
83.8% for C and 16.2% for G. The PCR results were confirmed by DNA
sequencing. The χ2 test showed that the genotype and
allele frequencies of PON2-148 do not deviate from
Hardy-Weinberg equilibrium (χ2=2.80, P>0.05).
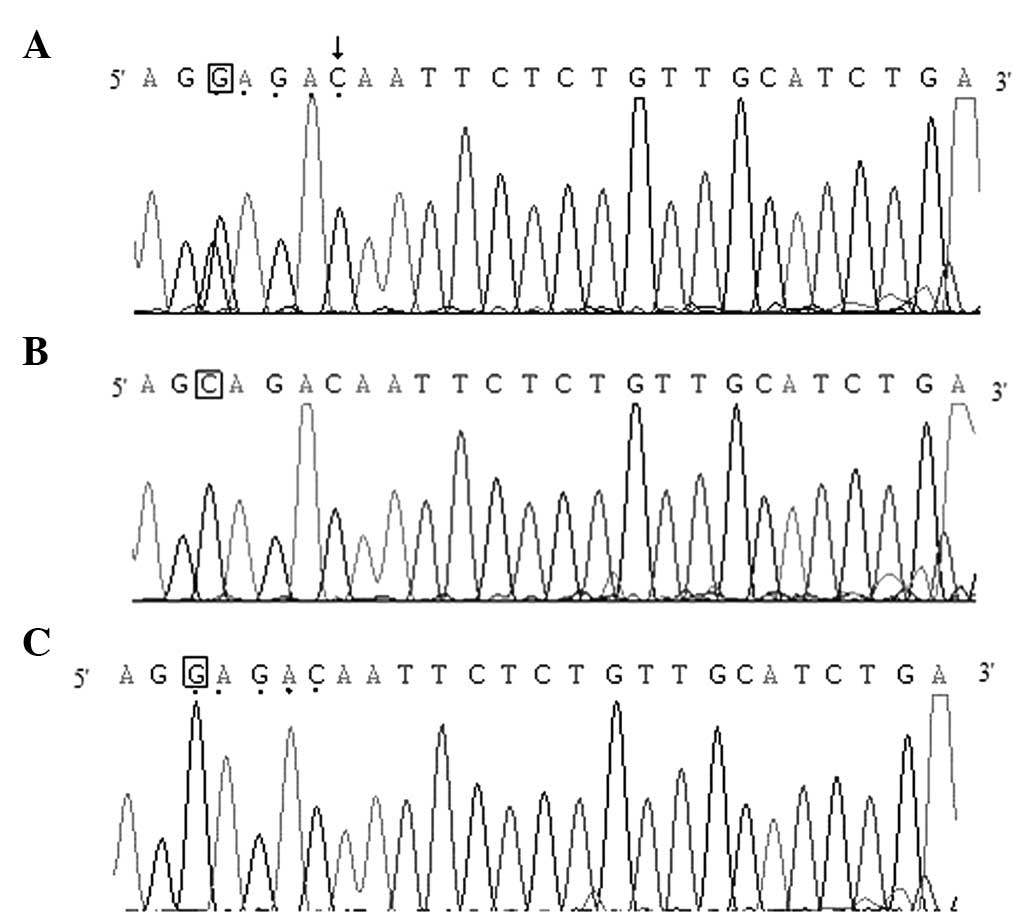
Examples of DNA sequencing of the PCR product of the
PON2 gene are shown in Fig. 2.
As expected, the sequences of the amplified products were
consistent with the one published in Genbank (NM_000446), with the
exception of the mismatched base.
The BsmAI recognition site (GAGAC) is
indicated by a marked symbol in Fig. 2A
and C, and the vertical arrow corresponds to the mismatched
base. The genotypes of GC, CC and GG are shown in Fig. 2, respectively, and the bases
representing the polymorphism sites are denoted in squares.
Discussion
The commonly used detection methods for SNP analysis
include DNA sequencing, TaqMan assay, fluorescently labeled probes,
molecular beacons and PCR-RFLP. DNA sequencing is an accurate
method for genotyping; however, it is labor-intensive and not
suitable for high-throughput analysis. The most prominent advantage
for the TaqMan probe technique is that it does not require a PCR
post-treatment process, such as separation or elution, which
improves the detection rate. To ensure accurate classification is
difficult, as the probe must be designed and can be expensive
(12). Molecular beacons are marked by
fluorescent dyes of different colors to achieve the simultaneous
detection of multiple SNPs. However, dyes are less for the suitable
fluorescent dye-labeled, which significantly limits the detection
of flux. In addition, a fluorescent molecular beacon probe must be
designed and these are expensive (13). The abovementioned methods do have their
own advantages; however, the equipment used is expensive and the
experiment methods are complex, therefore, they are difficult to
popularize and apply for SNP detection. The PCR-RFLP technique is
widely used in the detection of SNP as the detection method is
simple, and it has a high sensitivity and reliability. However,
certain assays can be employed to detect rs12026 polymorphism of
PON2 using common equipment. The PCR-RFLP method has been used
since 1997 (14), but the BsoFI
enzyme or its isoschizomers required are expensive. In order to
overcome this, PCR-RFLP with mismatched primers and the inexpensive
HinfI enzyme was developed to detect rs12026 (15). However, the polyacrylamide gel
electrophoresis in this developed method was inconvenient compared
with the agarose gel electrophoresis. The present study introduced
the inexpensive BsmAI enzyme and fast agarose gel
electrophoresis to detect PON2 polymorphism rs12026 using
the PCR-RFLP method. In conclusion, a simple and economical
technique for analysis of the PON2 polymorphism rs12026 was
reported. This may be used widely in the future study of the
genetics of diseases, including cardiovascular disease,
cerebrovascular disease and diabetes.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. NSFC81001239 and
30972457).
References
|
1
|
Mochizuki H, Scherer SW, Xi T, Nickle DC,
Majer M, Huizenga JJ, Tsui LC and Prochazka M: Human PON2 gene at
7q21.3: Cloning, multiple mRNA forms, and missense polymorphisms in
the coding sequence. Gene. 213:149–157. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shiner M, Fuhrman B and Aviram M: A
biphasic U-shape effect of cellular oxidative stress on the
macrophage anti-oxidant paraoxonase 2 (PON2) enzymatic activity.
Biochem Biophys Res Commun. 349:1094–1099. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ng CJ, Wadleigh DJ, Gangopadhyay A, Hama
S, Grijalva VR, Navab M, Fogelman AM and Reddy ST: Paraoxonase-2 is
a ubiquitously expressed protein with antioxidant properties and is
capable of preventing cell-mediated oxidative modification of low
density lipoprotein. J Biol Chem. 276:44444–44449. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saeed M, Perwaiz Iqbal M, Yousuf FA,
Perveen S, Shafiq M, Sajid J and Frossard PM: Interactions and
associations of paraoxonase gene cluster polymorphisms with
myocardial infarction in a Pakistani population. Clin Genet.
71:238–244. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thameem F, He X, Voruganti VS, Nath SD,
Fanti P, Blangero J, Maccluer JW, Comuzzie AG, Arar NH and Abboud
HE: Evaluation of polymorphisms in paraoxonase 2 (PON2) gene and
their association with cardiovascular-renal disease risk in Mexican
Americans. Kidney Blood Press Res. 32:200–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chi DS, Ling WH, Ma J, Xia M, Hou MJ, Wang
Q, Zhu HL, Tang ZH and Yu XP: Relationship between paraoxonase 1 55
Met/Leu, paraoxonase 2 148 Ala/Gly genetic polymorphisms and
coronary artery disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
23:289–293. 2006.(In Chinese). PubMed/NCBI
|
|
7
|
Pasdar A, Ross-Adams H, Cumming A, Cheung
J, Whalley L, St Clair D and MacLeod MJ: Paraoxonase gene
polymorphisms and haplotype analysis in a stroke population. BMC
Med Genet. 7:282006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calle R, McCarthy MI, Banerjee P, Zeggini
E, Cull CA, Thorne KI, Wiltshire S, Terra S, Meyer D, Richmond J,
et al: Paraoxonase 2 (PON2) polymorphisms and development of renal
dysfunction in type 2 diabetes: UKPDS 76. Diabetologia.
49:2892–2899. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinizzotto M, Castillo E, Fiaux M, Temler
E, Gaillard RC and Ruiz J: Paraoxonase2 polymorphisms are
associated with nephropathy in Type II diabetes. Diabetologia.
44:104–107. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen D, Hu Y, Chen C, Yang F, Fang Z, Wang
L and Li J: Polymorphisms of the paraoxonase gene and risk of
preterm delivery. Epidemiology. 15:466–470. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shin BS: Paraoxonase gene polymorphism in
south-western Korean population. J Korean Med Sci. 24:561–566.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Kok JB, Wiegerinck ET, Giesendorf BA
and Swinkels DW: Rapid genotyping of single nucleotide
polymorphisms using novel minor groove binding DNA oligonucleotides
(MGB probes). Hum Mutat. 19:554–559. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Steemers FJ, Ferguson JA and Walt DR:
Screening unlabeled DNA targets with randomly ordered fiber-optic
gene arrays. Nat Biotechnol. 18:91–94. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hegele RA, Connelly PW, Scherer SW, Hanley
AJ, Harris SB, Tsui LC and Zinman B: Paraoxonase-2 gene (PON2) G148
variant associated with elevated fasting plasma glucose in
noninsulin-dependent diabetes mellitus. J Clin Endocrinol Metab.
82:3373–3377. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zheng F, Du H, Krepinsky JC,
Segbo JA and Zhou X: Detecting the polymorphisms of paraoxonase
(PON) cluster in Chinese Han population based on a rapid method.
Clin Chim Acta. 365:98–103. 2006. View Article : Google Scholar : PubMed/NCBI
|