Introduction
Aortic dissection (AD) is the most common
life-threatening cardiovascular disorder, with an annual incidence
ranging from 5 to 30 per 100,000 individuals (1). This condition has a high mortality
(2); ~3% of the patients succumb
suddenly at the time of onset, and 50% within two days, and the
1-month mortality is as high as 90% in patients who do not undergo
surgery (3). With improvements in the
overall standard of living causing individuals to live longer, the
incidence of AD is increasing, thereby causing a decline in medical
resources and social wealth. A detailed investigation on the
molecular mechanism underlying AD may facilitate with delaying or
even reversing arterial wall reconstruction, consequently reducing
the incidence of AD and improving patient prognosis.
The pathogenesis of AD remains unclear; several
genes are considered to be involved in its occurrence. For example,
patients with Marfan syndrome suffer from deficiency of the
glycoprotein, fibrillin-1 (4), while
patients with Ehlers-Danlos syndrome have abnormal type-III
precollagen (5), such as deletions or
bona fide amino acid substitutions, which cause delayed formation
and destabilization of the collagen triple helix and, as a
consequence, reduced secretion of the molecule, which leads to
weakened arterial walls. Such patients may suffer from AD at a
particularly young age; however, the majority of patients with AD
do not exhibit such explicit syndromes. Thus, a detailed
investigation of the genes associated with the pathogenesis of AD
may help with the early identification and control of this disease
by modifying the genes, and via other methods prior to the onset of
AD.
In the present study, human whole genome microarray
was employed to screen differential genes from AD and normal aortic
tissue samples. In addition, reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) was adopted to verify the
downregulated genes, and the results were compared with the results
of microarray analysis in order to investigate the role of
differentially expressed genes screened by gene microarray in the
pathogenesis of AD.
Materials and methods
Tissue sampling
Five AD specimens were obtained from patients with
non-hereditary AD who were undergoing cardiovascular surgery in
Fuwai Hospital (Beijing, China) during the time period from July
2014 to December 2014. Patients with familial AD and those who had
aortic diseases caused by gene mutation were excluded. Four control
specimens were obtained from age-matched patients undergoing heart
surgery for non-aortic diseases. The fresh tissues were rinsed with
cold phosphate-buffered saline to remove the blood and thrombus on
the surface, quickly frozen in liquid nitrogen, and stored at −80°C
until use. The present study was approved by the medical ethics
committee of Fuwai Hospital and written informed consent was
obtained from all patients.
Total RNA extraction and quality
test
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
purified using the RNeasy mini kit (Qiagen China Co., Ltd.,
Shanghai, China), according to the manufacturer's instructions. The
purity of the total RNA was tested using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA) with 2 µl total RNA per sample, and integrity was tested using
an Agilent 2100 BioAnalyzer (Agilent Technologies, Inc., Santa
Clara, CA, USA).
Microarray hybridization, scanning and
analysis
Using RNA as templates, single-strand and
double-strand cDNAs were synthesized using the One-cycle cDNA
Synthesis kit (Affymetrix, Inc., Santa Clara, CA, USA), followed by
combining, washing and elution steps to purify the cDNA.
Thereafter, biotin-marked aRNA was in vitro transcribed
using an RNA transcription kit (Qiagen China Co., Ltd.) with
double-stranded cDNA as templates, followed by fragment processing.
Subsequently, the qualified cDNA was pre-hybridized with the
GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Inc.) at
45°C for 10 min using a Hybridization Oven 640 (Affymetrix, Inc.).
Subsequently, the hybridization solution was removed and an equal
volume of prepared hybridization solution was added for
hybridization at 45°C for 16 h. The microarray was then eluted and
stained using Fluidics Station 450, and was ultimately scanned
using the GeneChip® Scanner 3000 (Affymetrix, Inc.), after which
the scanned images and raw data were processed using the
Affymetrix® GeneChip® Command Console® operating system
(Affymetrix, Inc.).
Microarray data processing and
bioinformatics analysis
Microarray data were analyzed using GCOSvL.4
software according to the standard procedures provided by
Affymetrix, the principle and steps of which were referred from the
documentation ‘Genechip Expression Analysis-Data Analysis
Fundamentals’. The microarray data was analyzed using the Gene
Ontology (GO; http://www.geneontology.org/gene-associations/) and
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) databases. GO terms were
collected from the Gene Ontology and National Center for
Biotechnology Information (www.ncbi.nlm.nih.gov/gene/data/GO) databases. Based on
this analysis, the five AD specimens and four healthy aortic
specimens were orthogonally compared to determine the
differentially expressed genes. The ratio of fluorescence signal
intensity of AD to normal aortic tissue samples was calculated and
transformed to the base 2 logarithm, where a positive value was
referred to as an upregulated gene (≥±2 represented statistically
significant upregulation) and a negative value referred to a
downregulated gene (≤±2 represented statistically significant
downregulation).
Validation of expression profiles by
RT-qPCR
Six genes with significant (P<0.05) differential
expressions [myosin light chain kinase (MYLK), polycystin 1,
transient receptor potential channel interacting (PKD-1),
myosin heavy chain 11 (MYH11), superoxide dismutase 3,
extracellular (SOD3), filamin A (FLNA) and transgelin
(TAGLN)] were selected to verify the microarray results
using RT-qPCR. The microarray assay and RT-qPCR were performed
using the same batch of specimens. The RNAs were extracted using
TRIzol reagent, reverse transcribed using Moloney murine leukemia
virus (Takara Biotechnology Co., Ltd., Shiga, Japan), and
separately subjected to PCR amplification. The primers were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.) using
GAPDH as the reference gene. The primer sequences, amplification
length and annealing temperature are presented in Table I. The reaction procedure was as
follows: Initial denaturation at 94°C for 5 min; followed by 35
cycles for denaturation at 94°C for 30 sec, annealing for 30 sec
and elongation at 72°C for 30 sec; each sample was assayed in
triplicate. A temperature range of 65–95°C was selected for drawing
melting curves.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Target gene | Upstream primer
(5′-3′) | Downstream primer
(5′-3′) | Length of product
(bp) | Annealing temperature
(°C) |
|---|
| PKD-1 |
GGACCATCAACGACAAGCAG |
CCATCCCCAAAGTTCCACAG | 280 | 58 |
| MYH11 |
TGTGTCGTGGCTACTTG |
ATTCTCTGCCTTCTGCT | 233 | 52 |
| MYLK |
TCTCATTTACCATTCTGATGGC |
GCGCATTCAAAGCTTTTTTC | 169 | 55 |
| SOD3 |
TACCGAAACACCCCGCTCA |
TGCCAAACATTCCCCCAAA | 325 | 55 |
| FLNA |
ACGTGCTTCACCAGGATCTC |
CCTGGCAGCTACCTCATCTC | 353 | 58 |
| TAGLN |
ATGGCCAACAAGGGTCCATCC |
TCCATCTGCTTGAAGACCATG | 275 | 53 |
| GAPDH |
ATGGCCAACAAGGGTCCATCC |
TCCATCTGCTTGAAGACCATG | 275 | 55 |
Statistical analysis
Data were analyzed using SPSS 21.0 software (IBM
SPSS, Armonk, NY, USA). All measurement data are reported as the
mean ± standard deviation. Comparisons involving measurement data
and rates were performed between groups using paired Student's
t-tests and χ2 analysis, respectively. P<0.05 was
considered to indicate a statistically significant difference. As
per microarray analyses, the target gene expression was considered
to increase when the ratio of its expression in the AD
tissue/healthy aorta tissue was ≥2-fold, and was considered to
decrease when the ratio was ≤0.5.
Results
Quality testing of total RNA
The A260/A280 value was found to range from 1.8 to
2.2, the optical density (OD)260/OD230 value was found to range
from 1.5 to 2.0, and the RNA integrity index was >6.0. In
addition, the results demonstrated absence of genomic DNA
contamination and good RNA integrity; furthermore, the total RNA
28s/18s peak area ratio of the nine specimens was ~2:1, indicating
that the RNA accorded with the requirement for microarray
hybridization.
Screening of the differentially
expressed genes by gene expression microarray
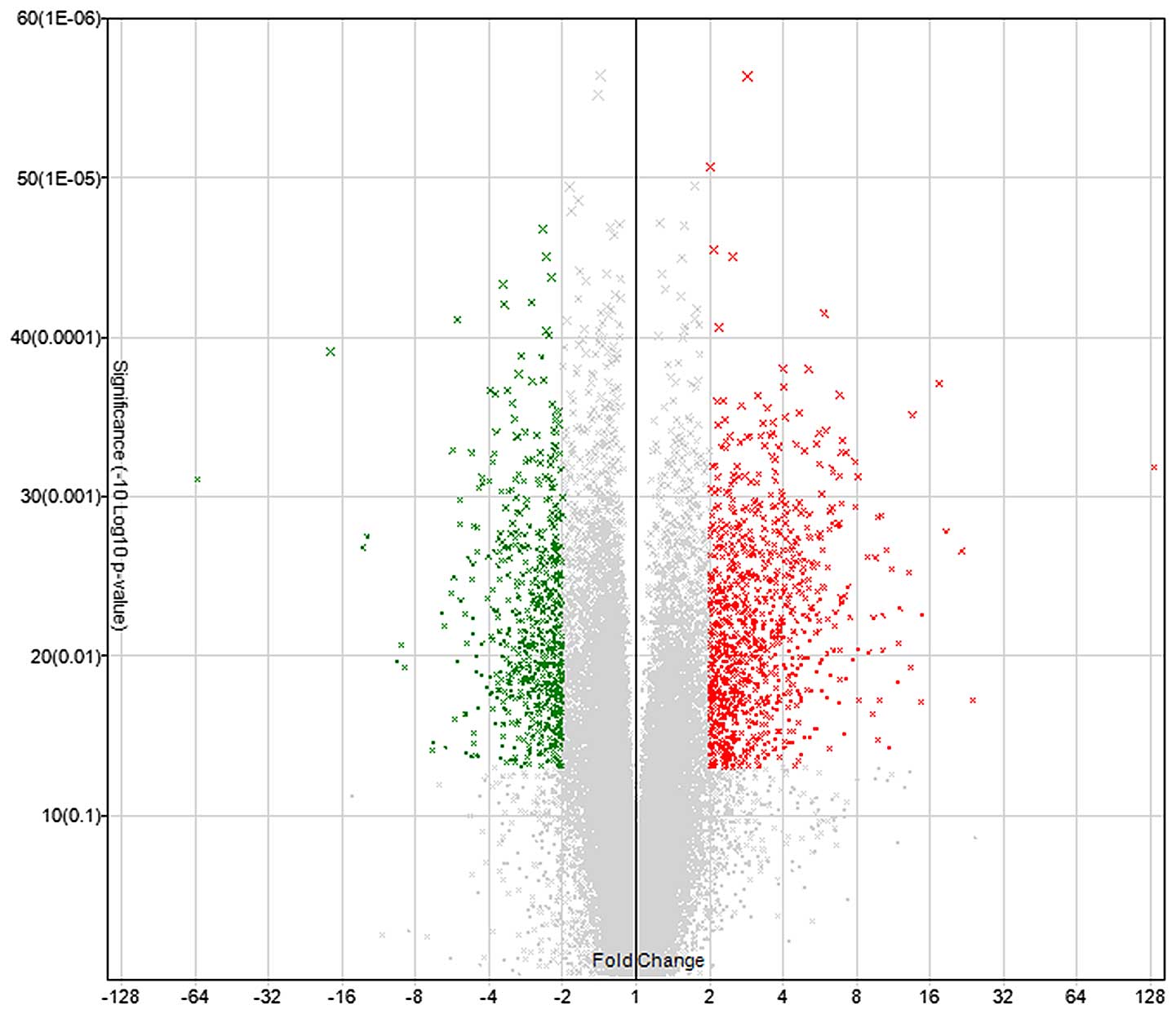
Microarray hybridization revealed that, compared
with normal aortic tissues, a total of 1,661 genes in the dissected
aortic tissues exhibited greater than 2-fold differential
expression (Fig. 1), of which 997
genes were upregulated and 664 genes were downregulated.
GO analyses of differential genes
The screened genes with greater than 2-fold
differential expression were subjected to GO analyses. These genes
were divided into three categories: Biological process, cellular
component and molecular function. The differential genes
predominantly involved in cellular immune response, inflammatory
reaction, intracellular signaling cascades, and regulation of cell
proliferation, cell death and apoptosis, and cell cycle were
classified in the category of biological process. The genes that
showed significant expression in the cytoplasmic vesicle, myosin
complex and extracellular matrix were assigned to the cellular
component category. The genes that were primarily involved in actin
binding, cytoskeletal protein binding, enzyme binding, calmodulin
binding, nucleoside binding, ATP binding, and activation of protein
dimer, GTPase, serine/serine protein kinase, and peptase were
assigned to the molecular function category (Table II).
 | Table II.GO functional analysis of
differentially expressed genes. |
Table II.
GO functional analysis of
differentially expressed genes.
| GO ID | GO term | Genes associated with
the GO term, n | P-value |
|---|
| GO:0006955 | Immune response | 125 | 5.67E-28 |
| GO:0009611 | Response to
wounding | 93 | 8.62E-20 |
| GO:0006954 | Inflammatory
response | 67 | 5.28E-18 |
| GO:0006952 | Defense response | 97 | 2.14E-17 |
| GO:0048584 | Positive regulation
of response to stimulus | 44 | 1.84E-10 |
| GO:0050778 | Positive regulation
of immune response | 33 | 2.75E-10 |
| GO:0007242 | Intracellular
signaling cascade | 135 | 2.79E-10 |
| GO:0045321 | Leukocyte
activation | 41 | 1.47E-08 |
| GO:0007155 | Cell adhesion | 82 | 4.57E-08 |
| GO:0042127 | Regulation of cell
proliferation | 89 | 5.75E-08 |
| GO:0010941 | Regulation of cell
death | 86 | 1.89E-06 |
| GO:0042981 | Regulation of
apoptosis | 85 | 2.04E-06 |
| GO:0007049 | Cell cycle | 78 | 3.50E-05 |
| GO:0001666 | Response to
hypoxia | 22 | 8.73E-05 |
| GO:0015629 | Actin
cytoskeleton | 48 | 1.01E-10 |
| GO:0044459 | Plasma membrane
part | 207 | 2.19E-10 |
| GO:0044421 | Extracellular
region part | 98 |
|
| GO:0003779 | Actin binding | 52 | 2.20E-10 |
| GO:0008092 | Cytoskeletal
protein binding | 67 | 1.23E-09 |
| GO:0005516 | Calmodulin
binding | 23 | 2.90E-05 |
| GO:0019865 | Immunoglobulin
binding |
7 | 1.42E-04 |
| GO:0005524 | ATP binding | 105 | 0.043192 |
| GO:0030695 | GTPase regulator
activity | 35 | 0.028338 |
| GO:0005088 | Ras
guanyl-nucleotide exchange factor activity | 11 | 0.037291 |
Biological pathways for the
differentially expressed genes
The central tendencies of the differentially
expressed genes in the pathways were determined by the P-value,
where a smaller P-value referred to a stronger central tendency.
The following signaling pathways were found to be associated with
the most differentially expressed genes within the threshold range:
FcγR-mediated phagocytosis, chemokine signaling pathway, complement
and coagulation cascade, leukocyte transendothelial migration and
cell adhesion molecule (Table
III).
 | Table III.Kyoto Encyclopedia of Genes and
Genomes biological pathways for differentially expressed genes. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes biological pathways for differentially expressed genes.
| Pathway | Count | P-value |
|---|
| FcγR-mediated
phagocytosis | 22 | 7.02E-06 |
| Chemokine signaling
pathway | 33 | 1.09E-05 |
| Complement and
coagulation cascades | 17 | 4.73E-05 |
| Viral
myocarditis | 17 | 6.86E-05 |
| Leukocyte
transendothelial migration | 22 | 2.08E-04 |
| Type I diabetes
mellitus | 12 | 2.33E-04 |
| Allograft
rejection | 11 | 2.66E-04 |
| Cell adhesion
molecules | 23 | 3.91E-04 |
| Natural killer cell
mediated cytotoxicity | 23 | 4.35E-04 |
| Systemic lupus
erythematosus | 19 | 4.48E-04 |
| B cell receptor
signaling pathway | 16 | 4.66E-04 |
| Graft vs. host
disease | 11 | 5.39E-04 |
| Regulation of actin
cytoskeleton | 30 | 0.001841243 |
| Intestinal immune
network for IgA production | 11 | 0.00348427 |
| Autoimmune thyroid
disease | 11 | 0.004719241 |
| Hematopoietic cell
lineage | 15 | 0.0054399 |
| Nucleotide-binding
oligomerization domain-like receptor signaling pathway | 12 | 0.006862366 |
| Cytokine-cytokine
receptor interaction | 32 | 0.009531305 |
| Toll-like receptor
signaling pathway | 16 | 0.009654688 |
| Dilated
cardiomyopathy | 15 | 0.009906976 |
| p53 signaling
pathway | 12 | 0.01367287 |
| Peroxisome
proliferator-activated receptors signaling pathway | 12 | 0.015182779 |
| Extracellular
matrix-receptor interaction | 13 | 0.025860442 |
| Hypertrophic
cardiomyopathy | 13 | 0.02810487 |
| Focal adhesion | 24 | 0.033743536 |
| FcγRI signaling
pathway | 12 | 0.034919203 |
| Amyotrophic lateral
sclerosis | 9 | 0.04755248 |
Verification of the differential genes
by RT-qPCR
In order to verify the reliability of microarray
results, the AD-associated genes, MYLK, MYH11,
SOD3, FLNA, PKD-1 and TAGLN were
screened according to their expression and GO classification using
RT-qPCR; the results are shown in Table
IV. Compared with the normal aortic tissue specimens, the mRNA
expression in MYLK, MYH11, SOD3, PKD-1
and TAGLN genes in the AD group was significantly decreased
(P<0.05), while that in FLNA was decreased, although the
difference was not statistically significant.
 | Table IV.Gene validation using RT-qPCR
(n=3). |
Table IV.
Gene validation using RT-qPCR
(n=3).
|
|
| Microarray | RT-qPCR |
|---|
|
|
|
|
|
|---|
| Gene | Molecular
function | Fold-change (mean ±
SD) | P-value | Fold-change (mean ±
SD) | P-value |
|---|
| MYLK | Actin binding,
cytoskeletal protein binding, calmodulin binding, protein kinase
activity, magnesium ion binding, purine ribonucleotide binding,
protein serine/threonine kinase activity, calcium ion binding,
adenyl nucleotide binding, ribonucleotide binding | −2.14±0.165 | 0.005 | 0.478±1.708 | 0.028 |
| MYH11 | Actin binding,
cytoskeletal protein binding, calmodulin binding, structural
constituent of muscle, protein kinase activity, magnesium ion
binding, adenyl nucleotide binding, ribonucleotide binding | −3.7±0.201 | 0.015 | 0.476±0.089 | 0.017 |
| FLNA | Actin binding,
cytoskeletal protein binding | −2.11±0.246 | 0.031 | 0.945±0.151 | 0.053 |
| SOD3 | Carbohydrate
binding, copper ion binding, polysaccharide binding, pattern
binding, heparin binding, polysaccharide binding | −2.08±0.150 | 0.003 | 0.380±0.032 | 0.001 |
| TAGLN | Actin binding,
cytoskeletal protein binding | −2.43±0.178 | 0.008 | 0.386±0.020 | 0.005 |
| PKD-1 | Protein complex
binding, sugar binding | −2.48±0.145 | 0.002 | 0.469±0.151 | 0.022 |
Discussion
Investigations into the pathogenesis of AD have long
been a major topic in academia. The pathogenesis of AD is
considered to be a complex multifactorial process that involves
several factors, including heredity (such as Marfan syndrome),
hypertension, atherosclerosis, trauma, inflammation, oxidative
stress and autoimmune diseases (6,7). However,
the mechanism by which these factors induce AD remains unclear.
According to modern molecular biology, various external factors
induce the occurrence and development of diseases by changing the
gene expression profiles of tissues or cells. Thus, analyzing these
changes enables us to understand the mechanism underlying the
occurrence and development of the diseases at the molecular level.
In the present study, the gene expression profiles of AD and normal
aorta tissues were analyzed using human whole-genome microarray,
and 1,661 genes with greater than 2-fold differential expression
were screened. These differential genes were involved in the
cellular immune response, intracellular signaling cascades, and
regulation of cell proliferation, cell death and apoptosis, as well
as inflammatory reactions and oxidative stress in the cell cycle
and other biological processes.
Inflammatory reactions occurring in the arterial
wall are important in the incidence of cardiovascular diseases,
including macrovascular atherosclerosis, myocardial infarction and
congestive heart failure (8–10). He et al (11) employed anti-cluster of differentiation
3 (CD3) and anti-CD45 to evaluate the expression of T-lymphocytes
in the aortic walls of patients with AD, and used anti-CD68 to
observe the expression of macrophages. Their results revealed that
compared with normal controls, elevated expression levels of CD3,
CD45 and CD68 were observed in the aortic walls of patients with
AD. In addition, macrophages are one of the earliest inflammatory
cells invading the aorta, and they are predominantly involved in
the reconstruction of the arterial wall by secreting
metalloproteinases, collagenase, elastase and pro-inflammatory
cytokines, which lead to aortic diseases. Yuan et al
(12) observed the gene and protein
levels of matrix metalloproteinase-2 (MMP-2) and MMP-9 in human AD
specimens and identified macrophage-mediated chronic inflammation
in the arterial wall. Tieu et al (13) developed an aortic reconstruction model
by treating ApoE−/− mice with angiotensin II,
and confirmed that interleukin (IL)-6 promoted the monocytes to
differentiate into macrophages and induce cells to generate CDl4
and CDllb, thereby increasing the expression levels of monocyte
chemoattractant protein-1 and MMP-9. All these findings indicate
that inflammatory factors are important in the incidence of AD.
However, in the present study, 67 inflammation-associated genes
exhibited significant differential expression in AD, of which IL-8,
IL-6, IL-10, CD14, CD163 and secreted phosphoprotein 1, among
others were significantly upregulated.
Previous studies have demonstrated that
cardiovascular diseases, such as atherosclerosis, hypertension,
atrial fibrillation, myocardial ischemia and cardiomyopathy are
closely associated with reactive oxygen species. Liao et al
(14) investigated the oxidative
stress-associated indicators in the AD and normal aortic walls, and
found that the malondialdehyde level in the AD group was
significantly higher than in the normal aorta group, while the
total SOD activity and genotype SOD activity were lower in the AD
group than those in the normal aorta group. This indicated that the
aortic wall of patients with AD was more severely attacked by free
radicals and possessed a weaker ability to clear the free radicals
when compared with the normal aortic wall, thereby suggesting
oxidative stress injury in AD (14).
The microarray results in the present study revealed that 22
oxidative stress-associated genes were significantly changed in the
AD tissue samples. In addition, microarray analysis and RT-qPCR
revealed that the expression of the SOD3 gene was
significantly decreased in the AD group, which is consistent with
previous results (14).
Similar results regarding genes that were
significantly upregulated or downregulated in the AD tissue samples
assessed in the present study have previously been reported, and
studies have shown that certain genes were involved in various
biological processes. For example, polycystic kidney disease
gene-1-encoded protein product, namely polycystin-l (PC-l),
participates in cell adhesion and cell signal transduction. Hassane
et al (15) conducted
experiments using a PKD-1 knockout mouse model, and found
that the downregulation of PKD-1 expression caused the mouse
aorta to generate potential space and intramural hematoma, and led
to a series of structural changes that are similar to those
occurring prior to AD. The microarray analysis and RT-qPCR
validation performed in the present study also revealed significant
downregulation of PKD-1 expression in patients with AD.
The medial layer of the aorta is predominantly
composed of vascular smooth muscle cells and ~45–55 layers of
elastin. Vascular smooth muscle cells are key in the maintenance of
vascular homeostasis, regulation of blood pressure, and damage
repair processes, and maintain the arterial tension through their
own contractile function, as well as by synthesizing and secreting
extracellular matrix (16). Thus,
smooth muscle cell proliferation and phenotype modulation
abnormalities are likely to alter the activities of extracellular
matrix and proteolytic enzymes, and promote the characteristic
structural diseases in the development of AD. Meanwhile,
MYLK encoding the myosin light-chain kinase (MLCK) protein
and the MYH11 gene are the genes encoding special myosins
for smooth muscle cells, which are the important components of
contractile units of smooth muscle cells (17,18).
Previous study reported that certain human chronic
and acute diseases are associated with increased MLCK expression
levels or activity (19–21); MLCK was found to affect the damage
caused by the inflammatory reaction by regulating the myosin light
chain. MLCK regulates the contraction of smooth muscle cells
(22) and is the key signaling pathway
involved in the regulation of smooth muscle cell contraction.
Furthermore, as a multifunctional regulator, MLCK has non-kinase
actin and myosin-binding activity, and it is important in smooth
muscle cell proliferation, migration and reconstruction of the
cytoskeleton (23). Additionally, an
MYH11 gene mutation was reported to alter the contraction of
smooth muscle cells (24). In the
present study, microarray analysis and RT-qPCR verified that the
expression levels of MYLK and MYH11 were
significantly downregulated in the AD tissue samples, and that they
may be involved in the occurrence and development of AD by
regulating the contraction of smooth muscle cells.
In addition, the current study predicted that
certain genes, such as TAGLN and FLNA, might be
associated with AD, although, to the best of our knowledge, their
role in the development of AD has not been thoroughly investigated.
Microarray analysis revealed that the expression levels of these
two genes were significantly downregulated; however, RT-qPCR
confirmed that FLNA was downregulated in the AD group
without statistical significance, compared with the normal
group.
In the present study, high-density whole genome
expression microarray was used to identify differentially expressed
genes between AD and healthy aorta tissue samples, as well as to
analyze the function, location, and other data regarding the
differential genes. The results revealed that 997 genes were
upregulated and 664 genes were significantly downregulated in the
pathogenesis of AD. In addition, six differential genes were
selected for verification by RT-qPCR, which indicated that the
expression levels of these six genes were downregulated in the AD
group compared with the healthy aorta group, five of which were
significantly downregulated (P<0.05) and that FLNA was
downregulated, although it was not statistically significant
(P>0.05). In conclusion, these findings demonstrate that the
genes screened by microarray were closely associated with the
pathogenesis of AD, providing a basis for further investigation of
the molecular mechanism underlying the pathogenesis of AD.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81270385).
References
|
1
|
Mészáros I, Mórocz J, Szlávi J, Schmidt J,
Tornóci L, Nagy L and Szép L: Epidemiology and clinicopathology of
aortic dissection. Chest. 117:1271–1278. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang S, Wang J, Lin P, Li Z, Yao C, Chang
G, Li X and Wang S: Short-term curative effect of endovascular
stent-graft treatment for aortic diseases in China: A systematic
review. PLoS One. 8:e710122013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gandet T, Canaud L, Ozdemir BA, Ziza V,
Demaria R, Albat B and Alric P: Factors favoring retrograde aortic
dissection after endovascular aortic arch repair. J Thorac
Cardiovasc Surg. 150:136–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang F, Li B, Lan L and Li L: C596G
mutation in FBN1 causes Marfan syndrome with exotropia in a Chinese
family. Mol Vis. 21:194–200. 2015.PubMed/NCBI
|
|
5
|
Reinstein E, DeLozier CD, Simon Z, Bannykh
S, Rimoin DL and Curry CJ: Ehlers-Danlos syndrome type VIII is
clinically heterogeneous disorder associated primarily with
periodontal disease, and variable connective tissue features. Eur J
Hum Genet. 21:233–236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scali ST, Waterman A, Feezor RJ, Martin
TD, Hess PJ Jr, Huber TS and Beck AW: Treatment of acute visceral
aortic pathology with fenestrated/branched endovascular repair in
high-surgical-risk patients. J Vasc Surg. 58:56–65.e1. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Faure EM, Canaud L, Agostini C, Shaub R,
Böge G, Marty-ané C and Alric P: Reintervention after thoracic
endovascular aortic repair of complicated aortic dissection. J Vasc
Surg. 59:327–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wales KM, Kavazos K, Nataatmadja M, Brooks
PR, Williams C and Russell FD: N-3 PUFAs protect against aortic
inflammation and oxidative stress in angiotensin II-infused
apolipoprotein E-/- mice. PLoS One. 9:e1128162014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brasier AR: The nuclear
factor-kappaB-interleukin-6 signalling pathway mediating vascular
inflammation. Cardiovasc Res. 86:211–218. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maiellaro K and Taylor WR: The role of the
adventitia in vascular inflammation. Cardiovasc Res. 75:640–648.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He R, Guo DC, Estrera AL, Safi HJ, Huynh
TT, Yin Z, Cao SN, Lin J, Kurian T, Buja LM, et al:
Characterization of the inflammatory and apoptotic cells in the
aortas of patients with ascending thoracic aortic aneurysms and
dissections. J Thorac Cardiovasc Surg. 131:671–678. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan Y, Wang C, Xu J, Tao J, Xu Z and
Huang S: BRG1 overexpression in smooth muscle cells promotes the
development of thoracic aortic dissection. BMC Cardiovasc Disord.
14:1442014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tieu BC, Lee C, Sun H, Lejeune W, Recinos
A III, Ju X, Spratt H, Guo DC, Milewicz D, Tilton RG, et al: An
adventitial IL-6/MCP1 amplification loop accelerates
macrophage-mediated vascular inflammation leading to aortic
dissection in mice. J Clin Invest. 119:3637–3651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liao M, Liu Z, Bao J, Zhao Z, Hu J, Feng
X, Feng R, Lu Q, Mei Z, Liu Y, et al: A proteomic study of the
aortic media in human thoracic aortic dissection: implication for
oxidative stress. J Thorac Cardiovasc Surg. 136:65–72, 72.e1-3.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hassane S, Claij N, Lantinga-van Leeuwen
IS, Van Munsteren JC, Van Lent N, Hanemaaijer R, Breuning MH,
Peters DJ and DeRuiter MC: Pathogenic sequence for dissecting
aneurysm formation in a hypomorphic polycystic kidney disease 1
mouse model. Arterioscler Thromb Vasc Biol. 27:2177–2183. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ito S, Ozawa K, Zhao J, Kyotani Y,
Nagayama K and Yoshizumi M: Olmesartan inhibits cultured rat aortic
smooth muscle cell death induced by cyclic mechanical stretch
through the inhibition of the c-Jun N-terminal kinase and p38
signaling pathways. J Pharmacol Sci. 127:69–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Guo DC, Cao J, Gong L, Kamm KE,
Regalado E, Li L, Shete S, He WQ, Zhu MS, et al: Mutations in
myosin light chain kinase cause familial aortic dissections. Am J
Hum Genet. 87:701–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bellini C, Wang S, Milewicz DM and
Humphrey JD: Myh11(R247C/R247C) mutations increase thoracic aorta
vulnerability to intramural damage despite a general biomechanical
adaptivity. J Biomech. 48:113–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen D, Lin Y and Xiong Y: Epithelial MLCK
and smooth muscle MLCK may play different roles in the development
of inflammatory bowel disease. Dig Dis Sci. 59:1068–1069. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Cheng Z, Qu X, Dai H, Ke X and
Chen Z: Overexpression of myosin is associated with the development
of uterine myoma. J Obstet Gynaecol Res. 40:2051–2057. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zou DB, Wei X, Hu RL, Yang XP, Zuo L,
Zhang SM, Zhu HQ, Zhou Q, Gui SY and Wang Y: Melatonin inhibits the
Migration of Colon Cancer RKO cells by Down-regulating Myosin Light
Chain Kinase Expression through Cross-talk with p38 MAPK. Asian Pac
J Cancer Prev. 16:5835–5842. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Milewicz DM, Guo DC, Tran-Fadulu V, Lafont
AL, Papke CL, Inamoto S, Kwartler CS and Pannu H: Genetic basis of
thoracic aortic aneurysms and dissections: Focus on smooth muscle
cell contractile dysfunction. Annu Rev Genomics Hum Genet.
9:283–302. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Betapudi V: Life without double-headed
non-muscle myosin II motor proteins. Front Chem. 2:452014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Renard M, Callewaert B, Baetens M, Campens
L, MacDermot K, Fryns JP, Bonduelle M, Dietz HC, Gaspar IM, Cavaco
D, et al: Novel MYH11 and ACTA2 mutations reveal a role for
enhanced TGFβ signaling in FTAAD. Int J Cardiol. 165:314–321. 2013.
View Article : Google Scholar : PubMed/NCBI
|