Introduction
Diabetes mellitus (DM) and hypertension (HTN) are
considered as major public health problems worldwide. The two
rarely exist in isolation. The World Health Organization has ranked
the Kingdom of Saudi Arabia (KSA) as having the seventh highest
rate in the world, and the second highest rate of diabetes in the
Middle East, with ~7 million individuals living with diabetes and
>3 million who are pre-diabetics (1). In KSA, DM is a rapidly increasing medical
health problem and is becoming a significant cause of morbidity
and/or mortality (2).
In addition to traditional life-style risk factors,
genetic background may also be pivotal in the pathogenesis of type
2 DM (T2DM) and HTN with the important and complex involvement of
the renin angiotensin aldosterone system (RAAS) (3).
Polymorphisms of the angiotensin-converting enzyme
(ACE) gene have been extensively investigated to ascertain the
genetic susceptibility of HTN development (4). In addition, ACE significantly contributes
to the pathogenesis of T2DM as RAAS blockade was demonstrated to
improve insulin resistance (5). A
meta-analysis reported the ACE insertion deletion (I/D)
polymorphism as a candidate gene for the development of essential
HTN in a Chinese population (6). Thus,
the I/D polymorphism of the ACE gene may significantly influence
the advancement of DM and/or HTN.
Different studies have described the link between
the D allele of the ACE gene and the development of HTN (7) and T2DM in various populations (8). However, certain studies failed to show
this association with the D allele (9). The contradictory results regarding the
involvement of the ACE I/D polymorphism in HTN and T2DM are likely
to be due to variance in ethnicity and/or gender (10,11). To
properly manage this issue in a Saudi Arabian population, a
multidisciplinary approach is required.
Yet, to date there is a paucity of information on
the association between the ACE I/D polymorphism and the risk of
T2DM in Saudi Arabian individuals. Therefore, the current study was
designed to establish the association of ACE I/D polymorphism in
Saudi Arabian subjects with T2DM and/or HTN.
Patients and methods
Ethical consideration and
participants
Ethical approval was obtained from the Ethics
Committee of the Faculty of Medicine, Prince Sattam Bin Abdelaziz
University (PSAU; Akharj, KSA). Participants included 220 subjects
that were attending the outpatient medicine Clinic at PSAU Hospital
over an 8-month period (January to August 2016). All participants
provided written informed consent prior to beginning the study.
Inclusion criteria and study
groups
Participants included subjects with T2DM and/or HTN.
T2DM was defined by the American Diabetes Association (ADA)
criteria as fasting blood glucose level >7.0 mmol/l (12). Subjects with persistent elevation of
blood pressure ≥140/90 mmHg or who were on antihypertensive
medications during the study period were considered hypertensive
and, therefore, included in the study. Healthy individuals were
randomly collected from volunteers and PSAU staff members without
age or sex matching. Their inclusion criteria were defined as
fasting blood glucose level <7.0 mmol/l, blood pressure
<140/90 mmHg, negative family history of diabetes and HTN, and
absence of any medication intake at the time of enrollment.
The study included four groups as follows: Control
group (n=48); T2DM group (n=70); HTN group (n=48); and T2DM with
HTN group (n=54). A questionnaire in Arabic and English language
was competed by each participant to obtain information regarding
socio-demographic status, history of diabetes, HTN and other
co-morbidities. Weight and height of all participants were
determined to compute their body mass index (BMI) using the
formula, weight (kg)/(height; m)2 (13).
Biochemical analysis of blood
samples
Blood samples were obtained from all subjects and
plasma was removed by centrifugation at 1,000 × g (4°C) for 10 min
and maintained at −30°C for subsequent analysis that included the
triglycerides (TG), high density lipoprotein cholesterol (HDL-C),
and total cholesterol (TC) levels. The Friedewald formula (14) was used to calculate low density
lipoprotein cholesterol (LDL-C). Lipid profile results were
classified according to the Third Report of the National
Cholesterol Education Program guidelines (15). The analysis was performed on a
Hitachi-912 auto-analyzer (Hitachi, Ltd., Tokyo, Japan) using kits
purchased from Roche Diagnostics GmbH (Mannheim, Germany).
Genotype determination
Saliva self-collection kits were used for genomic
DNA extraction according to manufacturer's instructions (DNA
Genotek, Inc., Kanata, ON, Canada). The samples were stored in a
freezer at −80°C until further analysis. The extracted DNA was
assessed for purity using a BioPhotometer (Eppendorf, Hamburg,
Germany). The ACE gene I/D polymorphism was determined by fragment
amplification using a polymerase chain reaction (PCR) technique
followed by gel electrophoresis. The flanking primer pairs were as
follows: Forward, 5′-GCCCTGCAGGTGTCTGCAGCATGT-3′ and reverse,
5′-GGATGGCTCTCCCCGCCTTGTCTC-3′ (synthesized by Research Biolabs Pte
Ltd., Singapore) (16). Fragment
amplification was performed with a 25-µl PCR master mix (New
England BioLabs, Inc., Ipswich, MA, USA), which contained 20 pmol
of each primer, 0.4 mmol/l deoxynucleotide triphosphate, 2 mmol/l
magnesium chloride, 1X Taq buffer and 1 unit of NEB Taq DNA
polymerase. The PCR was performed on a Thermocycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) with preliminary
denaturation for 5 min at 94°C, followed by cycles of denaturation
(94°C for 30 sec), annealing (58°C for 1 min) and extension (72°C
for 2 min). These cycles were repeated 30 times followed by a final
extension at 72°C for 5 min, the PCR amplicons were subsequently
stored at 4°C. The desired DNA fragments were separated by agarose
gel electrophoresis (Promega Corporation, Madison, WI, USA)
performed in original electrophoresis tank (Elchrom Scientific AG,
Cham, Switzerland). The PCR product (7 µl) and 5 µl of loading dye
were mixed then loaded to 1% ethidium bromide gel. After 60 min at
a 120-V current, the gel was removed and visualized using a
ChemiDoc™ MP Gel imaging System (Bio-Rad Laboratories, Inc., USA)
under UV light. The initial PCR results revealed three genotypes: A
490-bp band (II), a 190-bp band (DD) and the two bands (ID).
ID heterozygotes may be mistyped as DD
homozygotes
Therefore, to augment the specificity of the DD
homozygotes, another PCR amplification was performed with an
insertion-specific primer pair used in each DD sample as follows:
Forward, 5′-TGGGACCACAGCGCCCGCCACTAC-3′ and reverse,
5′-TCGCCAGCCCTCCCATGCCC-ATAA-3′. The PCR conditions were adjusted
with an initial denaturation step at 94°C for 1 min, followed by 30
cycles of denaturation (94°C for 30 sec), annealing (67°C for 45
sec) and extension (72°C for 2 min). The second PCR product
exhibited an I allele amplicon (335 bp) or no products in the DD
allele homozygous samples. Genotyping was repeated twice on
different occasions with similar results obtained.
Statistical analysis
Statistical analyses were conducted using SPSS
(version 20.0; IBM Corp., Armonk, NY, USA). The descriptive data
and laboratory results were tested for Gaussian distributions by
Kolmogorov-Smirnov test and reported as means ± standard deviation.
Differences in descriptive data were analyzed by one-way analysis
of variance followed by Bonferroni post hoc multiple comparisons.
Genotype and allelic frequencies in all groups were reported as
numbers and percentage. The genotype data were examined for
aberration from expected by Hardy-Weinberg equilibrium and the
significant difference between groups was calculated using the
χ2 test of independence and/or Fisher's exact test.
Furthermore, the odds ratios (ORs) and 95% confidence intervals
(CIs) were calculated by multiple logistic regression analysis.
P<0.05 was considered to indicate a statistically significant
difference at 95% CI.
Results
Table I demonstrates
that significant differences exist in BMI between the control group
and the other investigated groups (P<0.05); however, when
compared with each other, BMI was not significantly different
between the T2DM, HTN and T2DM + HTN groups. The variables,
systolic blood pressure (SBP) and diastolic BP (DBP) at the time of
participation were significantly higher in the HTN groups compared
with the control group (P<0.001), but was not significantly
different between the patient groups (P>0.05). Regarding fasting
blood glucose levels, significant differences were observed in all
patient groups compared with the control group. HDL-C was
significantly increased in the T2DM groups (P<0.05), although
not in the HTN group. By contrast, no statistical differences in
LDL-C, TG and TC levels were identified in any of the study
groups.
 | Table I.Demographic and biochemical results of
the study groups. |
Table I.
Demographic and biochemical results of
the study groups.
| Parameter | Control group
(n=48) | Diabetic group T2DM
(n=70) | Hypertensive group
HTN (n=48) | HTN + T2DM group
(n=54) |
|---|
| Gender (M/F) | 28/20 | 28/42 | 22/26 | 24/30 |
| Age (years) | 32.2±10.9 | 42.9±12.5 | 39.8±13.9 |
49.8±10.4a,b |
| Body mass index
(kg/m2) | 24.5±6.2 | 37.7±7.9a | 38.7±7.8a | 37.8±8.2a |
| Fasting blood glucose
(mmol/l) | 4.7±0.8 | 9.7±1.2a | 7.5±0.6a | 10.7±3.4a,b |
| Systolic blood
pressure (mmHg) | 126.1±8.7 | 140±20a | 158±18a | 154±17a |
| Diastolic blood
pressure (mmHg) | 78.1±7.2 | 87±11a | 96±12a | 94±14a |
| Total cholesterol
(mmol/l) | 5.21±1.33 | 6.12±1.55 | 5.76±1.62 | 5.99±1.42 |
| Low density
lipoprotein-cholesterol (mmol/l) | 3.80±1.33 | 4.24±1.47 | 4.16±1.54 | 4.44±1.26 |
| High density
lipoprotein-cholesterol (mmol/l) | 0.90±0.45 |
1.77±0.26a | 1.08±0.31 |
1.67±0.39a |
| Triglycerides
(mmol/l) | 1.97±1.07 | 2.81±1.08 | 2.29±1.02 | 2.77±1.09 |
The ACE gene I/D polymorphism genotype frequencies
followed the Hardy-Weinberg equilibrium in all of the studied
groups (Table II). The control group
exhibited higher frequencies of the II and ID genotypes than the DD
genotype (58.3, 37.5 vs. 4.2% respectively). Compared with the
control group, all diabetic-associated groups exhibited a lower
frequency of II genotype than the ID and DD genotypes that was
statistically significant (χ2=48.83; P<0.0001). The
frequencies of the II genotype were 8.6, 4.2, and 5.6% in T2DM,
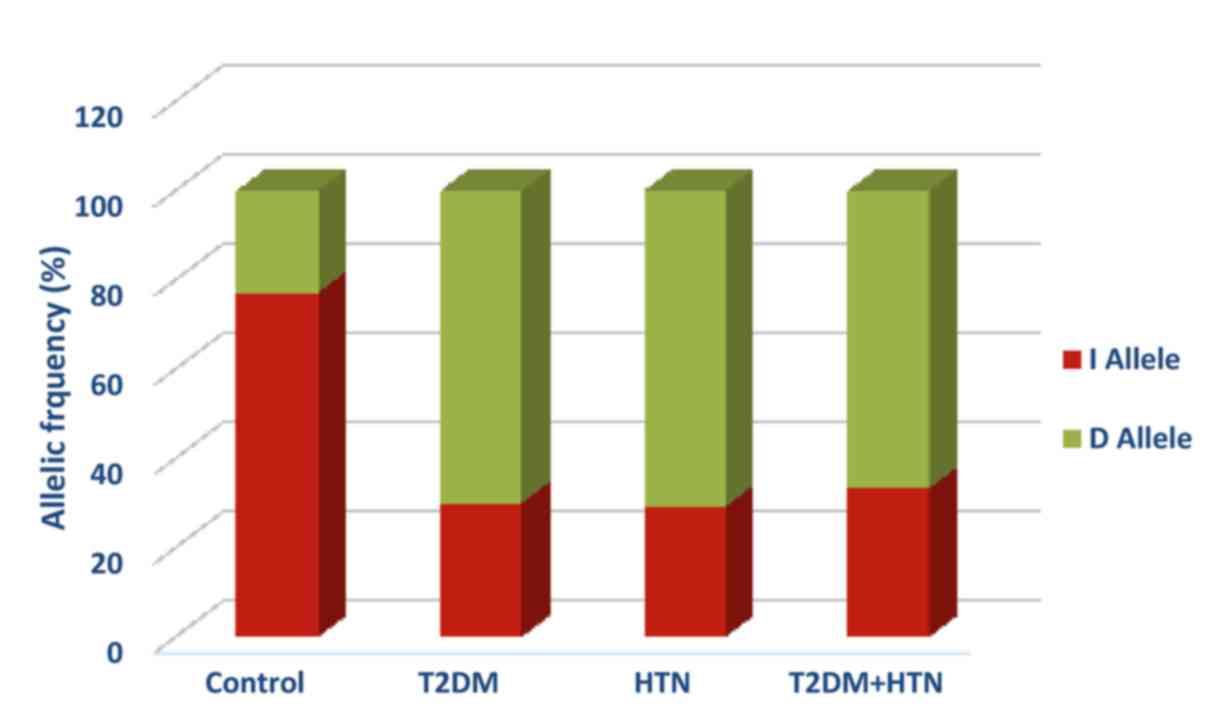
HTN, and T2DM with HTN, respectively (Table II). In addition, the D allele was the
dominant allele in diabetic, hypertensive, and
hypertensive-diabetic patients with frequencies of 70, 70.8, and
66.7% respectively (Fig. 1). The D
allele was significantly associated with T2DM diabetes (OR=7.85;
95% CI=3.90–15.78; P<0.0001), HTN (OR = 8.17; 95% CI = 3.73 –
17.88; p< 0.0001), and HTN-T2DM diabetes (OR=6.73; 95%
CI=2.90–15.59; P<0.0001) compared with the control group
(Table II). No significant
differences were observed between D allele and other
diabetes-associated co-morbidities.
 | Table II.I/D polymorphism genotyping and
allelic frequencies between the study groups. |
Table II.
I/D polymorphism genotyping and
allelic frequencies between the study groups.
| Parameter | Control group
(n=48) | T2DM (n=70) | HTN (n=48) | HTN + T2DM group
(n=54) |
|---|
| Genotype, n (%) |
|
|
|
|
| II | 28 (58.3) | 6 (8.6) | 2 (4.2) | 3 (5.6) |
| ID | 18 (37.5) | 30 (42.8) | 24 (50.0) | 30 (55.5) |
| DD | 2 (4.2) | 34 (48.6) | 22 (45.8) | 21 (38.9) |
| χ2 test
sig. (2-sided) |
| P<0.0001 between
all groups |
|
|
| Allele, n (%) |
|
|
|
| I | 74 (77.1) | 42 (30) | 28 (29.2) | 36 (33.3) |
| D | 22 (22.9) | 98 (70) | 68 (70.8) | 72 (66.7) |
|
Total | 96 | 140 | 96 | 108 |
| Fisher's exact test
sig. (2-sided) |
|
P<0.0001a |
P<0.0001a |
P<0.0001a |
| Odds ratio |
| 7.85 | 8.17 | 6.73 |
| (95% confidence
interval) |
| (3.90–15.78) | (3.73–17.88) | (2.90–15.59) |
Discussion
It has previously been reported that an increased
serum ACE level is genetically identified by a 287-bp fragment of
the I/D polymorphism at chromosome 17 in the 16th intron of the ACE
gene (17). The presence/insertion of
this fragment is demonstrated as homozygosis II, ID for
heterozygosis, while DD symbolizes the absence/deletion of a 287-bp
Alu reduplicate series. DD genotypes, primarily the D allele, in
contrast to the II genotypes and the I allele, are strongly
associated with an increased risk of diabetes and diabetic
complications (18), myocardial
infarction (19) and HTN (20). By contrast, many studies denied the
associated between the DD genotypes, and development of HTN
(9) and diabetes (21). Furthermore, the I allele had a strong
link with familial HTN in an Australian population (20).
Due to the inconsistent results of the
above-mentioned studies, the ACE gene polymorphism was investigated
in a Saudi Arabian T2DM patients with or without HTN in the present
study. A strong link between the DD homozygous genotype and,
primarily, the D allele of the ACE gene with T2DM and HTN was
identified in Saudi Arabian subjects, when compared with the
healthy control group (P<0.05; Table
II). Previous studies in other populations showed high
prevalence of the DD genotype (93.33%) and D allele (81.39%) of the
ACE gene in diabetic and HTN patients as compared with control
subjectss (7,21). In the present study the frequency of
the D allele was found to be significantly different in
hypertensive and T2DM patients with or without HTN compared with
the control subjects (P<0.0001), a similar result was reported
in a Taiwanese population (22).
In a Malaysian population, the angiotensinogen gene
polymorphism was reported to be linked with HTN subjects, which
clearly demonstrated an important role of the RAAS polymorphism in
the development of HTN (23). However,
another study denied the association of the renin gene and HTN,
with or without T2DM (24).
Data from a study that involved Malaysian
hypertensive subjects and angiotensinogen gene polymorphisms
support the current results with regard to the diabetic and
hypertensive groups' clinical characteristics and the ACE genotype
distribution in terms of age, fasting blood glucose, SBP, DBP, and
BMI, as the results are comparable (25). Additionally, the results of the present
study are consistent with other studies regarding T2DM subjects in
Taiwanese (26) and Iranian
populations (27); however, there was
negative association with gender, ethnicity and other confounding
factors. Numerous factors may contribute to explaining these
discrepancies that deny the link between the ACE I/D polymorphism,
and T2DM and HTN, including ethnic differences or heterogeneity of
the population, sampling bias, and potentially other ecological
factors.
In addition, there were certain limitations of the
present study; first, due to the controlled design, which was
randomly performed, it did not allow for age- and sex-matched
controls to be involved. Furthermore, the case subjects were
relatively older than the control individuals. However, despite the
somewhat small sample size of the current study compared with other
epidemiological and association studies, the results confirm the
assumption that the DD genotype, chiefly the D allele, is strongly
associated with T2DM and/or HTN. In order to emphasize the
association of the ACE I/D polymorphism with T2DM and HTN, further
research with larger sample sizes is essential and further
investigations are required to evaluate the possible correlation of
other polymorphisms of RAAS genes with the risk of T2DM and HTN
developing in a Saudi Arabian population.
In conclusion, the current study demonstrated strong
evidence for the association of ACE gene I/D polymorphisms and the
risk of T2DM and HTN in Saudi Arabian subjects. Furthermore, the D
allele of the ACE I/D polymorphism was demonstrated as a valuable
genetic marker for T2DM and HTN in Saudi Arabian subjects.
Acknowledgements
The authors would like to thank the Scientific
Research Deanship of Prince Sattam Bin Abdulaziz University
(Al-kharj, Saudi Arabia; grant no. 2015/03/4581), for providing
financial support. The authors would also like to thank Dr. Taimour
Langaee, Research Associate Professor, Department of
Pharmacotherapy and Translational Research at the University of
Florida (UF) College of Pharmacy and Director of the UF Center for
Pharmacogenomics, for his kind invitation to Dr Rehab Ashour and
supervision of the genotyping laboratory work.
References
|
1
|
Robert AA, Al Dawish MA, Braham R,
Musallam MA, Al Hayek AA and Al Kahtany NH: Type 2 Diabetes
Mellitus in Saudi Arabia: Major Challenges and Possible Solutions.
Curr Diabetes Rev. 13:59–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Al Dawish MA, Robert AA, Braham R, Al
Hayek AA, Al Saeed A, Ahmed RA and Al Sabaan FS: Diabetes Mellitus
in Saudi Arabia: A Review of the Recent Literature. Curr Diabetes
Rev. 12:359–368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramachandran V, Ismail P, Stanslas J,
Shamsudin N, Moin S and Jas R Mohd: Association of
insertion/deletion polymorphism of angiotensin-converting enzyme
gene with essential hypertension and type 2 diabetes mellitus in
Malaysian subjects. J Renin Angiotensin Aldosterone Syst.
9:208–214. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu X, Cshang YP, Yan D, Weder A, Cooper
R, Luke A, Kan D and Chakravarti A: Associations between
hypertension and genes in the renin angiotensin system.
Hypertension. 41:1027–1034. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dahm KA Jandeleit, Tikellis C, Reid CM,
Johnston CI and Cooper ME: Why blockade of the renin angiotensin
system reduces the incidence of new onset diabetes. J Hypertens.
23:463–473. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qu H, Lu Y and Lin S: Meta analysis on the
association of ACE/ID polymorphism and essential hypertension in
Chinese population. Zhonghua Yu Fang Yi Xue Za Zhi. 35:408–411.
2001.(In Chinese). PubMed/NCBI
|
|
7
|
Jeng JR, Harn HJ, Jeng CY, Yueh KC and
Shieh SM: Angiotensin I converting enzyme gene polymorphism in
Chinese patients with hypertension. Am J Hypertens. 10:558–561.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kennon B, Petrie JR, Small M and Connell
JM: Angiotensin converting enzyme gene and diabetes mellitus.
Diabet Med. 16:448–558. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiang FT, Lai ZP, Chern TH, Tseng CD, Hsu
KL, Lo HM and Tseng YZ: Lack of association of the angiotensin
converting enzyme polymorphism with essential hypertension in a
Chinese population. Am J Hypertens. 10:197–201. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barley J, Blackwood A, Cartere ND, Crews
DE, Cruickshank JK, Jeffery S, Ogunlesi AO and Sagnella GA:
Angiotensin converting enzyme insertion/deletion polymorphism:
association with ethnic origin. J Hypertens. 12:955–957. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Donell CJ, Lindpaintner K, Larson MG,
Rao VS, Ordovas JM, Schaefer EJ, Myers RH and Levy D: Evidence for
association and genetic linkage of the angiotensin converting
enzyme locus with hypertension and blood pressure in men but not
women in the Framingham Heart Study. Circulation. 97:1766–1772.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
American Diabetes Association, . Standards
of Medical Care in Diabetes-2016 Abridged for Primary Care
Providers. Clinical Diabetes: A Publication of the American
Diabetes Association. 34:3–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
WHO expert consultation. Appropriate body
mass index for Asian populations and its implications for policy
and intervention strategies. Lancet. 157:1632004.
|
|
14
|
Tremblay AJ, Morrissette H, Gagné JM,
Bergeron J, Gagné C and Couture P: Validation of the Friedewald
formula for the determination of low density lipoprotein
cholesterol compared with β quantification in a large population.
Clin Biochem. 37:785–790. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Expert panel on detection, evaluation, and
treatment of high blood cholesterol in adults. Executive summary of
the third report of the National Cholesterol Education Program
(NCEP) expert panel on detection, evaluation, and treatment of high
blood cholesterol in adults (Adult Treatment Panel III). JAMA.
285:2486–2497. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knoell DL, Johnston JS, Bao S and Kelley
KA: A genotyping exercise for pharmacogenetics in pharmacy
practice. Am J Pharm Educ. 73:432009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rigat B, Hubert C, Gelas F Alhenc, Cambien
F, Corvol P and Soubrier F: An insertion/deletion polymorphism in
the angiotensin I converting enzyme gene accounting for half the
variance of serum enzyme levels. J Clin Invest. 86:1343–1346. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marre M, Bernadet P, Gallois Y, Savagner
F, Guyene TT, Hallab M, Cambien F, Passa P and Gelas F Alhenc:
Relationship between angiotensin I converting enzyme gene
polymorphism, plasma levels, and diabetic retinal and renal
complication. Diabetes. 43:384–388. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Samani N, Thompson JR, O'Toole L, Channer
K and Woods KL: A meta analysis of the association of the deletion
allele of the angiotensin converting enzyme gene with myocardial
infarction. Circulation. 94:708–712. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zee RY, Lou YK, Griffiths LR and Morris
BJ: Association of a polymorphism of the angiotensin I-converting
enzyme gene with essential hypertension. Biochem Biophys Res
Commun. 184:9–15. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Daimon M, Oizumi T, Saitoh T, kameda W,
hirata A, Yamaguchi H, Ohnuma H, igarashi M, Tominaga M and Takeo
KA: The D allele of the angiotensin converting enzyme
insertion/deletion (I/D) polymorphism is a risk factor for type 2
diabetes in a population based Japanese sample. Endocr J.
50:393–398. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsieh MC, Lin SR, Hsieh TJ, Hsu CH, Chen
HC, Shin SJ and Tsai JH: Increased frequency of angiotensin
converting enzyme DD genotype in patients with type 2 diabetes in
Taiwan. Nephrol Dial Transplant. 15:1008–1013. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Say YH, Ling KH, Duraisamy G, Isaac S and
Rosli R: Angiotensinogen M235T gene variants and its association
with essential hypertension and plasma renin activity in Malaysian
subjects: A case control study. BMC Cardiovasc Disord. 5:72005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vasudevan R, Patimah I, Johnson S and
Norashikin S: No association of BglI dimorphism of human renin gene
in hypertensive subjects in Malaysia. Res J Biol Sci. 3:1218–1822.
2008.
|
|
25
|
Ghazali DM, Rehman A and Rahman AR:
Candidate gene polymorphisms and their association with
hypertension in Malays. Clin Chim Acta. 388:46–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tseng CH, Tseng CP, Chong CK, Sheu JJ and
Cheng JC: Angiotensin converting enzyme gene polymorphism and
stroke in type 2 diabetic patients in Taiwan. Eur J Clin Invest.
37:483–491. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nikzamir A, Nakhjavani M, Golmohamadi T
and Dibai L: Association of angiotensin-converting enzyme gene
insertion/deletion polymorphism with metabolic syndrome in Iranians
with type 2 diabetes mellitus. Arch Iran Med. 11:3–9.
2008.PubMed/NCBI
|