Introduction
Pathogenic fungi cause a wide range of damage in
organisms, including in plants, humans and other animals. To
protect themselves against fungal pathogens, living organisms
produce a myriad molecules. Different classes of antifungal
proteins isolated from various plants include chitinases,
cyclophilins (CyPs), defensins, lectins and lipid transfer proteins
(1–7),
all of which kill or suppress the infection of pathogenic
microorganisms. In addition, the introduction of genes encoding
these proteins into crop species has been found to confer enhanced
resistance on the resulting transgenic lines (8).
CyPs, also known as immunophilins, peptidyl-prolyl
cis-trans isomerases (PPIases) and cyclosporine A-binding
proteins, are expressed in a variety of organisms (plants, yeast,
fruit flies, parasites, rats and humans) (9) and exhibit high homology to one another.
In plants, CyPs were first reported in 1990 with the isolation of
CyP cDNA sequences from tomato, maize and oilseed rape (10). CyPs have endogenous PPIase activity
that isomerizes the cis-trans conformation of imide linkage in
substrates (11). Multiple CyP members
in plants such as rice and Arabidopsis are associated with
diverse functions and regulatory pathways related to their foldase,
chaperoning, scaffolding and other (unknown) activities (12–15).
Antifungal and antiviral activities of CyPs can relieve the
multiple stresses exerted by fungi and viruses (16). CyP-like antifungal proteins have been
isolated from black-eyed pea, mung bean, Chinese cabbage and
chickpea (4,17,18). The CyP
of Chinese cabbage has been shown to have pronounced effects on a
variety of fungal pathogens (17).
Plant CyPs have two isoforms that differ according
to the number of domains. The first isoform possesses only a single
PPIase domain, whereas the second type is composed of a catalytic
PPIase domain plus either a leucine zipper domain at the amino end,
a tetratricopeptide repeat domain at carboxyl end or another domain
related to sub-cellular localization (19). Members of the subfamily comprising
divergent CyPs have another loop containing the consensus sequence
XXGKXLH, a conserved Glu and two invariable Cys residues (20). It was reported that the divergent loop
can mediate protein-to-protein interactions or may be part of a
P-loop or ATP-binding site formed by residues 42-GEKCIGKS-49 and
163-VVIAD-167 (21).
Panax ginseng is a Chinese traditional herb
that is believed to have medicinal restorative properties (22). During growth, ginseng is exposed to
various soil-borne pathogenic microorganisms, including fungi,
bacteria and nematodes. However, the manner in which ginseng
resists fungi, especially through its protein contents remains to
be investigated. From ginseng transcriptome databases previously
established (23), a Panax
ginseng cyclophilin (pgCyP) of interest was identified since it
was highly induced during the period in which plant is highly
blight-prone. This observation suggested that pgCyP is involved in
the anti-microorganism process.
On the basis of that finding, the pgCyP gene
was cloned in the present study and expressed in a bacterial host.
We tagged pgCyP with 6×His to its end to facilitate chromatography.
We also enhanced its expression amount by applying the pGEX vector
with glutathione S-transferase (GST). The recombinant pgCyP
exhibited strong antifungal activity against Phytophthora
cactorum and also possessed PPIase activity.
Materials and methods
Biological material
Five-year-old plants of ginseng (Panax
ginseng C.A. Meyer) were harvested from Fusong County (Jinlin,
China). The freshly collected material was prepared for gene
cloning.
Phytopathogenic fungal species used in this study
were Rhizoctonia solani and Cylindrocarpon
destructans (Hyphomycetes); Phytophthora cactorum
(Oomycetes); Fusarium solani, Alternaria panax and
Botrytis cinerea (Fungi imperfecti); and Sclerotinia
sp. (Discomycetes). All the fungal species were obtained from
Jilin Agricultural University (Changchun, China).
Plasmid constructs
Total RNA was seperated from ginseng leaves. The
pgCyP gene was obtained by carrying out polymerase chain
reaction (PCR) amplification from leaf cDNA with synthetic
nucleotide primers. The PCR product was inserted into pMD18-T
(Takara, Dalian, China). After digestion with BamHI and
NotI, the generated DNA fragment was cloned in a pGEX-6p1
vector.
Expression of recombinant pgCyP
The recombinant plasmid was transformed into
Escherichia coli BL21 (DE3) to express pgCyP
His6. Transformed cells were cultured in Luria-Bertani
medium supplemented with 50 µg/ml ampicillin at 37°C on a rotary
shaker at 200 rpm. When the OD600 value of the cell
culture reached 0.6–0.8, protein overexpression was induced by the
addition of isopropyl β-D-1-thiogalactopyranoside (IPTG)
(Sigma-Aldrich, St. Louis, MO, USA) to a final concentration of 0.5
mM, and the culture was grown for a further 5 h. Cells were
harvested by centrifugation at 10,000 × g for 2 min. Protein
expression levels were analyzed by SDS-PAGE and visualized with
Coomassie Blue staining (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Isolation and purification of
recombinant pgCyP
The transformed cell pellets were resuspended in
wash buffer [20 mM Tris (pH 8.0), 100 mM NaCl and 3 M urea]. After
centrifugation, the cells were lysed in lysis buffer [50 mM
Tris-HCl, 1 mM EDTA, 100 mM NaCl (pH 8.0), 0.13 mM
phenylmethylsulfonyl fluoride, 0.5 mg/ml lysozyme and 1.33 mg/ml
sodium deoxycholate] and sonicated at 4°C for 30 min. DNase I was
then added to a concentration of 2,000 U/ml and the solution was
incubated at 37°C with shaking at 200 rpm for 1 h. The lysate was
then centrifuged at 10,000 × g for 20 min at 4°C. The resulting
inclusion bodies were washed twice with a solution consisting of 50
mM Tris, 100 mM NaCl, 2 M urea, 0.5% Triton-X and 10 mM EDTA at pH
8.0. The washed inclusion bodies were dissolved by stirring for 1 h
in extraction buffer [1.5% sarkosyl, 25 mM triethanolamine amine
and 1 mM EDTA (pH 8.0)] at 4°C. The solubilized inclusion bodies
were centrifuged at 10,000 × g for 10 min at 4°C and then refolded
by dialysis in binding buffer [10 mM
NaH2PO4·2H2O, 10 mM
Na2HPO4·12H2O (pH 7.8), 150 mM
NaCl and 10 mM imidazole]. The refolded protein was purified by
Ni-chelating Sepharose Fast Flow chromatography (GE Healthcare
Bio-Sciences AB, Uppsala, Sweden). The adsorbed proteins were
eluted with elution buffer containing 100 mM imidazole. Eluates
were pooled together and the samples were renatured on a Sephadex
G-25 column. Protein concentrations were determined by the Bradford
Protein Assay kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing,
China).
PPIase activity assay
A mixture of the following components was incubated
on ice for 10 min: 930 µl Assay Buffer [50 mM HEPES and 100 mM NaCl
(pH 7.8)], 30 µl of 200 µM α-chymotrypsin (Sigma-Aldrich) and
either 10 µl of GST-pgCyP-His6 (1 µM) or the GST (1 µM)
negative control. Each sample was placed in a spectrophotometer
(Thermo Fisher Scientific, Waltham, ΜΑ, USA), which was pre-cooled
to 8°C. After the addition of 30 µl of 7.8 mM
Suc-Ala-Ala-Pro-Phe-NA (Sigma-Aldrich), absorbance at 390 nm was
immediately recorded every second for 5 min at 8°C.
pgCyP antifungal activity
GST-pgCyP-His6 and GST protein were
tested for possession of antifungal activity. Fungi were grown in
potato dextrose agar (PDA) for 48 h at 28°C. The fungi were then
spread onto PDA plates. Sterilized blank paper disks were placed on
the plates and dotted with an aliquot of protein at different
concentrations. The plates were incubated and monitored for up to 3
days. To determine the IC50 of proteins against various
fungal pathogens, the fungal spores were collected and placed in
96-well microtiter plates. Recombinant pgCyP (20 µl) or the
negative control was then added to each well. After 12–36 h of
incubation at 28°C, fungal growth was evaluated microscopically.
The turbidity of each well was also measured by recording
absorbance at 595 nm using a microtiter reader (Emax, Molecular
Devices, Sunnyvale, CA, USA).
Sequence accession number
The nucleotide sequence of pgCyP in the present
study has an accession no. KX034081 in GenBank.
Results
Cloning and sequence analysis of pgCyP
in ginseng
As shown in Fig. 1A
(lane 3), pgCyP cDNA was successfully generated by reverse
transcription-polymerase chain reaction (RT-PCR) amplification from
cDNA of ginseng leaves. The corresponding nucleotide sequence,
consisting of 525 bp, was predicted to be 174 a.a. in length, as
well as a theoretical isoelectric point of 7.67 and a molecular
weight of 18.7 kDa. As it lacked a transit peptide, this predicted
protein was probably localized in the cytosol (24). The pgCyP protein contained a PPIase
domain and a divergent loop (48-VSGKPLH-54). Two CyPs (positioned
at 40 and 168) as well as a Glu (positioned at 83) were
conservatively kept similar to the other members in the CyPs
family. The predicted secondary structure of pgCyP comprised 6.32%
α helices, 21.26% extended strands and 72.42% random coils. On the
basis of protein sequence alignment, pgCyP was closely associated
with Ziziphus jujuba CyP, Citrus sinensis CyP19-3 and
Ricinus communis CyP proteins. pgCyP shares 90% identity
with its homolog in Ziziphus jujuba (Fig. 1D).
 | Figure 1.Detection of the pgCyP gene
transcript and sequence analysis. (A) PCR amplification of pgCyP by
RT-PCR from total mRNA of ginseng leaves. Marker refers to the DNA
molecular marker, while control is the mock-DNA negative control.
(B) Schematic representation of constructs of
GST-pgCyP-His6. (C) Predicted 3D structure model of
pgCyP. The 3D structure model was predicted using the SWISS-MODEL
server. (D) Alignment of the amino acid sequences of CyPs of
Panax ginseng and other species. Residues comprising the
divergent loop are shown in a black box, whereas the conserved Cys
residues (Cys-40 and Cys-168) and Glu (Glu-83) are indicated with
black arrows. pgCyP, Panax ginseng cyclophilin; PCR,
polymerase chain reaction; GST, glutathione S-transferase; RT-PCR,
reverse transcription-polymerase chain reaction; CyP, cyclophilin;
ca, Chinese cabbage; cs, Citrus sinensis; zj,
Ziziphus jujuba; rc, Ricinus communis. |
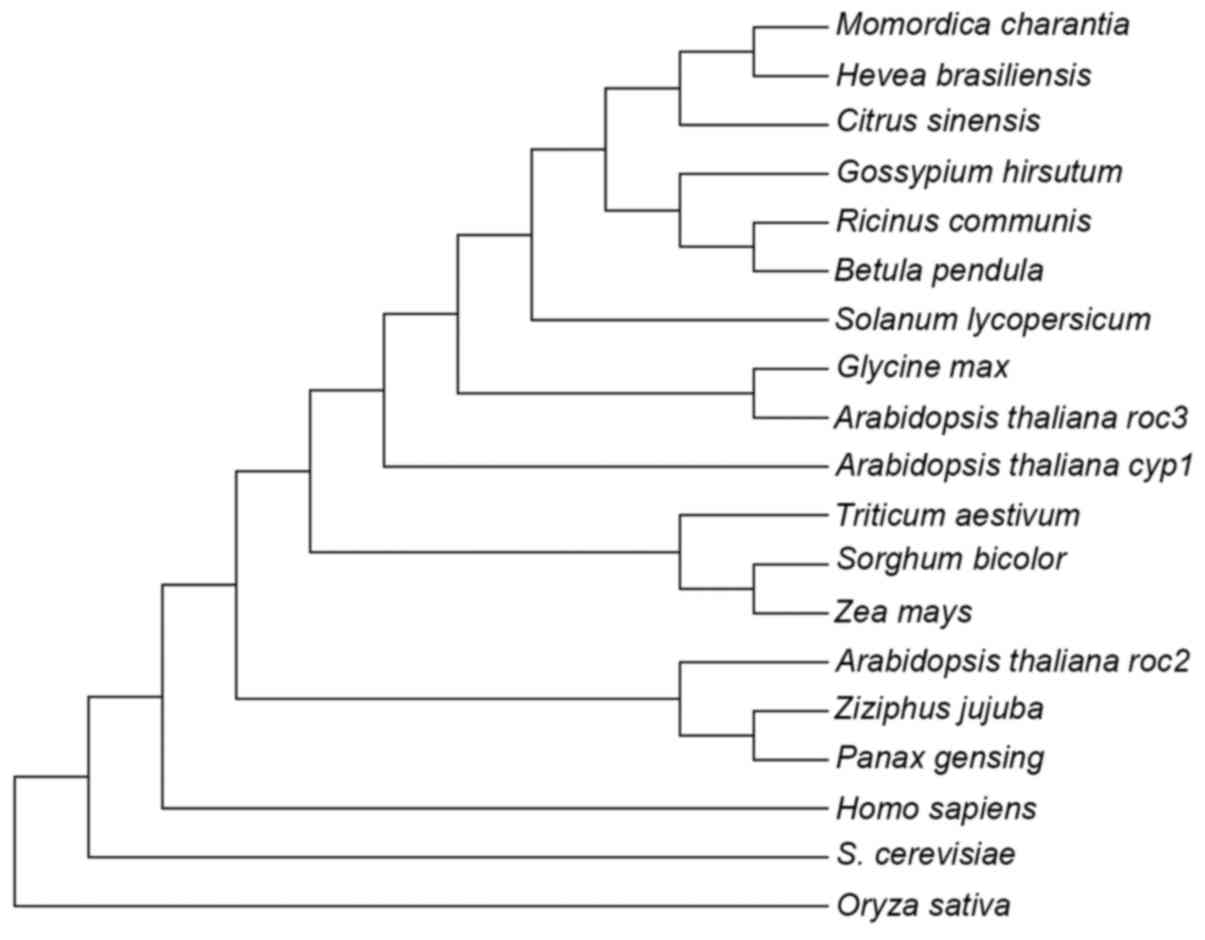
In the phylogenetic tree of CyP-like proteins shown
in Fig. 2, pgCyP may be classified, as
expected, into the clade of CyP proteins from dicot species, where
it is most closely related to Ziziphus jujuba belonging to
the same family. A 3D pgCyP model was created through primary
protein sequence (25) using the
SWISS-MODEL server (Fig. 1C).
Construction, expression and
purification of GST-pgCyP-His6
cDNA of the pgCyP isolated from this study, which
differs from the pgCyP previously reported (23), and possibly belongs to a different
ginseng CyP subfamily, was cloned into vector pMD18-T. The
resulting plasmid was then digested with BamHI and
NotI. After target fragments were cloned into pGEX-6p1, the
protein were carrying a tandem His tag and GST at both ends
(Fig. 1B).
Escherichia coli BL21 (DE3) competent cells
were transformed with the pGEX-6p1/pgCyP-His6
plasmid. Overexpression of recombinant protein was induced at
different IPTG concentrations for different lengths of time. The
optimal IPTG concentration and induction time was 0.5 mM and 5 h,
respectively. The induced protein was resolved in SDS-PAGE. As
expected, a protein bank of 46 kDa was evident (Fig. 3). Moreover, the analysis indicated
GST-pgCyP-His6 present as inclusion bodies (Fig. 3).
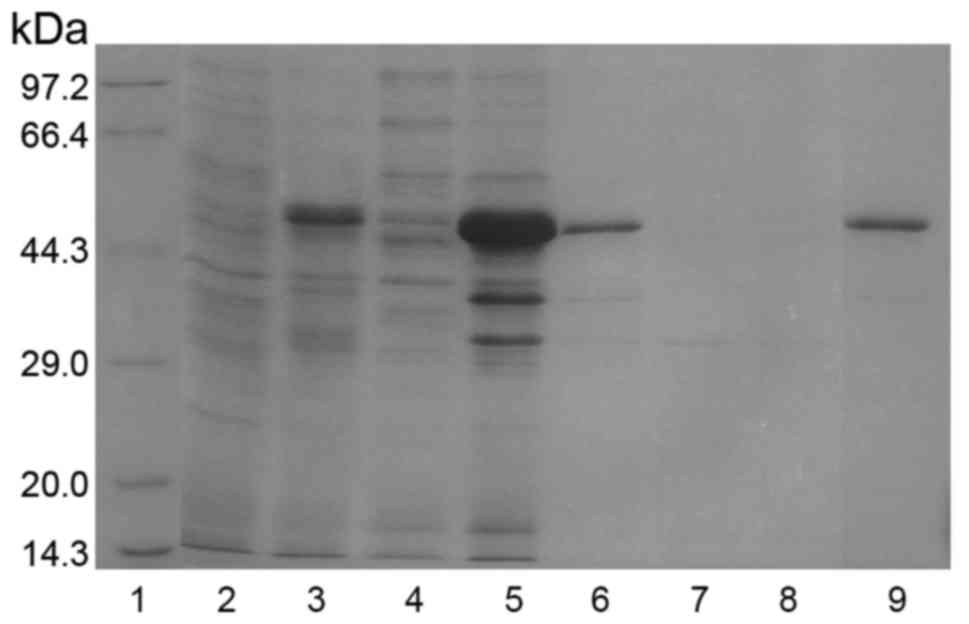 | Figure 3.Expression, purification and
refolding of GST-pgCyP-His6. Samples collected after
purification of GST-pgCyP-His6 were analyzed by sodium
dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie
Brilliant Blue staining. Lane 1, marker; lane 2, sample before IPTG
induction; lane 3, sample subjected to IPTG induction; lane 4,
soluble-protein fraction; lane 5, insoluble-protein fraction; lane
6, solubilized denatured insoluble-protein fraction; lane 7,
purification flow-through fraction; lane 8, purification wash
fraction; lane 9, purification elution fraction with 100 mM
imidazole. GST, glutathione S-transferase; pgCyP, Panax
ginseng cyclophilin; IPTG, isopropyl
β-D-1-thiogalactopyranoside. |
After solubilizing the inclusion bodies in
extraction buffer, pgCyP was purified using Ni-NTA affinity
chromatography as described in Materials and methods. The target
protein with His tag was fractionated with 100 mM imidazole.
GST-pgCyP-His6 was confirmed in SDS-Page (lane 9,
Fig. 3).
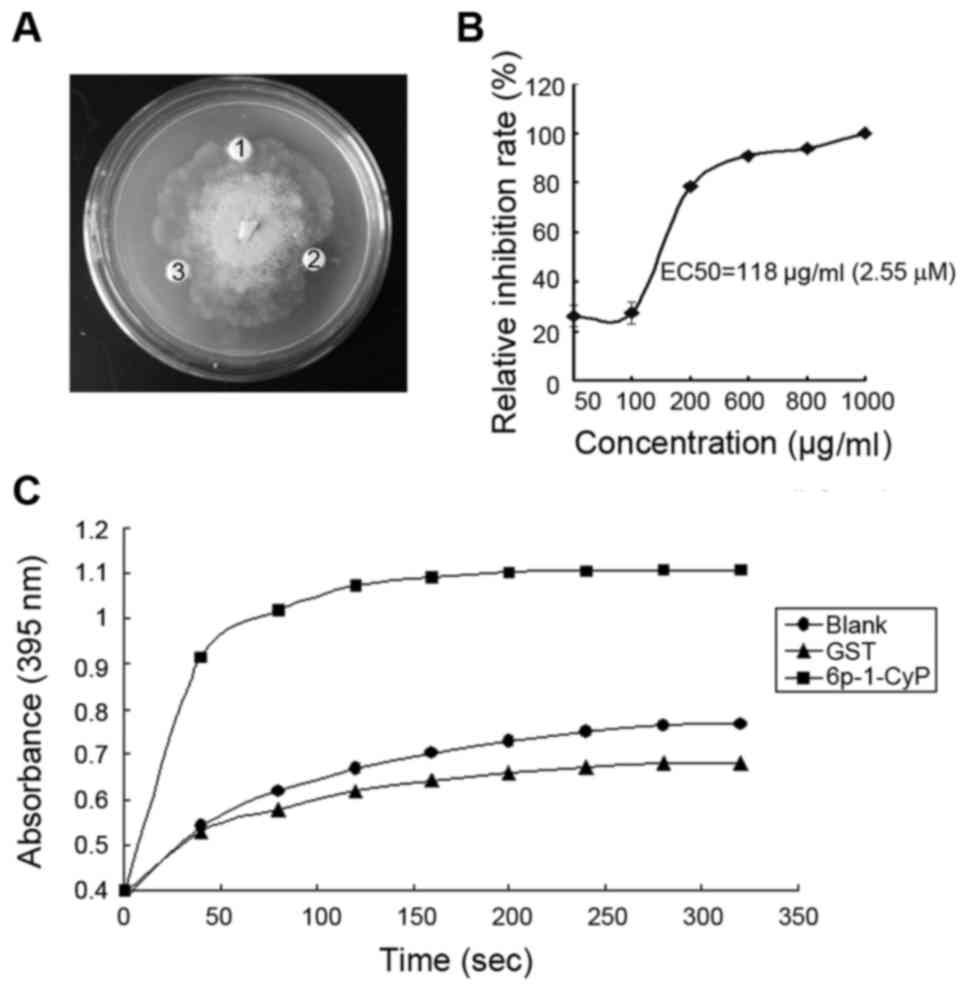
In vitro antifungal activity of
pgCyP
After purification and renaturation, the recombinant
protein was tested for its ability to inhibit fungal growth in
vitro. Pathogenic fungus included Sclerotinia sp., R.
solani, P. cactorum, F. solani, C.
destructans, A. panax and B. cinerea. The fusion
protein at concentrations of 1.28 and 2.14 µM had pronounced
effects on the growth of P. cactorum (Fig. 4A), but had no effect on the growth of
the other fungi. The IC50 of pgCyP against P.
cactorum, was 2.55 µΜ (Fig. 4B).
GST, as a negative control, was inactive. Hereby we demonstrate
in vitro resistance activity against fungi of pgCyP.
PPIase activity of recombinant
pgCyP
CyPs are PPIases
In the present study, we showed pgCyP has PPIase
activity. PgCyP is capable of accelerating isomerization of imide
between the Ala and Pro peptide bonds in contrast to spontaneous
inter-conversion in the negative control. GST protein, the control,
had no isomerize activity.
Discussion
Plant diseases are a major concern in the production
of agricultural crops and medicinal herbs. Although an increasing
number of antimicrobial peptides have been isolated from plants,
there is little research on anti-microorganism proteins of ginseng.
In the present study, we cloned and isolated a ginseng CyP protein.
PgCyP contains an ORF of 525 bp encoding 174 a.a. Ginseng blight is
very prevalent during the rainy season (July to September), and our
transcription expression level analysis revealed that CyP has a
high expression in roots during the same period. This finding
suggests a relationship between pgCyP expression and ginseng
blight. In the antifungal test, pgCyP exhibited strong antifungal
activity at micromolar concentrations against P.
cactorum.
The expression vectors PET26b, PET28a and pRSET B
were used; however, they failed to express pgCyP. GST, a chaperone
for protein folding, is frequently selected to help isolation of
soluble protein (26). However, use of
GST-fusion system for insoluble GST-fusion protein isolation
remains challenging (27). Ni-NTA
affinity chromatography was effective, and the one-step method was
used to isolate target proteins with His tag. Using this method, we
were able to obtain pure GST-fused protein. We then attempted to
hydrolyze the GST domain. Following protease treatment, the incised
protein was unstable and rapidly degraded. Consequently, the GST
tag was retained while performing the antifungal activity
tests.
The functional properties of the generated fusion
protein suggest pgCyP has PPIase activity. Taken together, our
results provide evidence that pgCyP has antifungal activity.
Previous findings have indicated that the CyP protein of Chinese
cabbage has antifungal activity against B. cinerea, T.
harzianum, T. viride, R. solani, F. solani
and F. oxysporum (17). In the
present study, however, pgCyP affected P. cactorum, growth
only, with no activity observed against R. solani, F.
solani or B. cinerea. pgCyP shares 78% identity with its
homolog in Chinese cabbage. Thus, the divergent pattern of
antifungal activity observed between the two homologous proteins
may be related to differences in amino acid sequences. We aim to
investigate the antifungal spectrum of pgCyP in a future study.
In conclusion, in this study, we carried out the
successful heterologous expression, purification and
characterization of pgCyP and investigated its structure and
function. While CyPs are reported to be involved in biotic stress
response, their exact functions remain to be identified. To
demonstrate the possible molecular functions executed,
identification of its downstream substrates is needed. Transgenic
plants expressing the pgCyP gene may facilitate revealing
the physiological functions in the future.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81373937, 81503212 and
81503324) and Jilin Scientific and Technological Development
Program (no. 20140520042JH).
Glossary
Abbreviations
Abbreviations:
|
CyP
|
cyclophilin
|
|
PPIase
|
peptidyl-prolyl cis-trans
isomerase
|
|
GST
|
glutathione S-transferase
|
|
PDA
|
potato dextrose agar
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
IPTG
|
isopropyl
β-D-1-thiogalactopyranoside
|
References
|
1
|
Kawase T, Yokokawa S, Saito A, Fujii T,
Nikaidou N, Miyashita K and Watanabe T: Comparison of enzymatic and
antifungal properties between family 18 and 19 chitinases from
S. coelicolor A3(2). Biosci Biotechnol Biochem. 70:988–998.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sattayasai N, Sudmoon R, Nuchadomrong S,
Chaveerach A, Kuehnle AR, Mudalige-Jayawickrama RG and
Bunyatratchata W: Dendrobium findleyanum agglutinin:
Production, localization, anti-fungal activity and gene
characterization. Plant Cell Rep. 28:1243–1252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wong JH and Ng TB: Sesquin, a potent
defensin-like antimicrobial peptide from ground beans with
inhibitory activities toward tumor cells and HIV-1 reverse
transcriptase. Peptides. 26:1120–1126. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ye XY and Ng TB: Isolation of unguilin, a
cyclophilin-like protein with anti-mitogenic, antiviral, and
antifungal activities, from black-eyed pea. J Protein Chem.
20:353–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin P, Xia L and Ng TB: First isolation of
an antifungal lipid transfer peptide from seeds of a
Brassica species. Peptides. 28:1514–1519. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang SY, Gong YS and Zhou JJ:
Chromatographic isolation and characterization of a novel
peroxidase from large lima legumes. J Food Sci. 74:C193–C198. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pelegrini PB, Noronha EF, Muniz MA,
Vasconcelos IM, Chiarello MD, Oliveira JT and Franco OL: An
antifungal peptide from passion fruit (Passiflora edulis)
seeds with similarities to 2S albumin proteins. Biochim Biophys
Acta. 1764:1141–1146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fritig B, Heitz T and Legrand M:
Antimicrobial proteins in induced plant defense. Curr Opin Immunol.
10:16–22. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Galat A: Variations of sequences and amino
acid compositions of proteins that sustain their biological
functions: An analysis of the cyclophilin family of proteins. Arch
Biochem Biophys. 371:149–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gasser CS, Gunning DA, Budelier KA and
Brown SM: Structure and expression of cytosolic
cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants
and production of active tomato cyclophilin in Escherichia
coli. Proc Natl Acad Sci USA. 87:pp. 9519–9523. 1990,
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fischer G, Bang H and Mech C:
Determination of enzymatic catalysis for the
cis-trans-isomerization of peptide binding in proline-containing
peptides. Biomed Biochim Acta. 43:1101–1111. 1984.(In German).
PubMed/NCBI
|
|
12
|
Kumari S, Roy S, Singh P, Singla-Pareek SL
and Pareek A: Cyclophilins: Proteins in search of function. Plant
Signal Behav. 8:e227342013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu A, He Z, Cho HS, Lima A, Buchanan BB
and Luan S: A chloroplast cyclophilin functions in the assembly and
maintenance of photosystem II in Arabidopsis thaliana. Proc
Natl Acad Sci USA. 104:pp. 15947–15952. 2007, View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahn JC, Kim DW, You YN, Seok MS, Park JM,
Hwang H, Kim BG, Luan S, Park HS and Cho HS: Classification of rice
(Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs,
CYPs) and expression patterns under water stress. BMC Plant Biol.
10:2532010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumari S, Singh P, Singla-Pareek SL and
Pareek A: Heterologous expression of a salinity and developmentally
regulated rice cyclophilin gene (OsCyp2) in E. coli and
S. cerevisiae confers tolerance towards multiple abiotic
stresses. Mol Biotechnol. 42:195–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong JH, Ng TB, Cheung RC, Ye XJ, Wang HX,
Lam SK, Lin P, Chan YS, Fang EF, Ngai PH, et al: Proteins with
antifungal properties and other medicinal applications from plants
and mushrooms. Appl Microbiol Biotechnol. 87:1221–1235. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JR, Park SC, Kim JY, Lee SS, Park Y,
Cheong GW, Hahm KS and Lee SY: Molecular and functional
characterization of a cyclophilin with antifungal activity from
Chinese cabbage. Biochem Biophys Res Commun. 353:672–678. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye XY and Ng TB: Isolation of a new
cyclophilin-like protein from chickpeas with mitogenic, antifungal
and anti-HIV-1 reverse transcriptase activities. Life Sci.
70:1129–1138. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Romano PG, Horton P and Gray JE: The
Arabidopsis cyclophilin gene family. Plant Physiol.
134:1268–1282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peterson MR, Hall DR, Berriman M, Nunes
JA, Leonard GA, Fairlamb AH and Hunter WN: The three-dimensional
structure of a Plasmodium falciparum cyclophilin in complex
with the potent anti-malarial cyclosporin A. J Mol Biol.
298:123–133. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dornan J, Page AP, Taylor P, Wu S, Winter
AD, Husi H and Walkinshaw MD: Biochemical and structural
characterization of a divergent loop cyclophilin from
Caenorhabditis elegans. J Biol Chem. 274:34877–34883. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arushanian EB and Beĭer EV: Participation
of pineal gland in antistressor activity of adaptogenic drugs. Eksp
Klin Farmakol. 78:9–12. 2015.(In Russian). PubMed/NCBI
|
|
23
|
Liu J, Wang Q, Sun M, Zhu L, Yang M and
Zhao Y: Selection of reference genes for quantitative real-time PCR
normalization in Panax ginseng at different stages of growth
and in different organs. PLoS One. 9:e1121772014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang P and Heitman J: The cyclophilins.
Genome Biol. 6:2262005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Campos BM, Sforça ML, Ambrosio AL,
Domingues MN, de Souza Brasil TA, Barbosa JA, Paes Leme AF, Perez
CA, Whittaker SB, Murakami MT, et al: A redox 2-Cys mechanism
regulates the catalytic activity of divergent cyclophilins. Plant
Physiol. 162:1311–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harper S and Speicher DW: Purification of
proteins fused to glutathione S-transferase. Methods Mol Biol.
681:259–280. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park DW, Kim SS, Nam MK, Kim GY, Kim J and
Rhim H: Improved recovery of active GST-fusion proteins from
insoluble aggregates: Solubilization and purification conditions
using PKM2 and HtrA2 as model proteins. BMB Rep. 44:279–284. 2011.
View Article : Google Scholar : PubMed/NCBI
|