Introduction
Human endogenous retroviruses (HERVs) belonging to
the superfamily of transposable and retrotransposable genetic
elements represent ~8% of the human genome (1). HERV type K (HERV-K) is the sole group of
endogenous retroviruses that is established to contain
human-specific members. Group HERV-K (also known as human mouse
mammary tumour virus like-2, HML-2) occupies ~5% of the DNA created
by insertions of human-specific transposable elements and is among
the most studied groups of human retroelements (2). The HERV-K group may be divided into 10
families. The designation ‘K’ originates from their use of a lysine
transfer RNA to prime reverse transcription, while ‘HML-2’
indicates the relationship to the murine betaretrovirus mouse
mammary tumour virus (2).
The disease association of HERVs has been a focus of
research, and there have been a number of studies on the different
expression profiles of HERVs in various clinical situations:
According to certain studies, HERVs may serve a significant role in
embryonic development, thereby contributing to formation of the
placenta, and may have correlation with cancer and autoimmune
diseases (3–6). However, there are few studies on the
transpositions of HERVs in the human genome. Furthermore, to the
best of our knowledge, there have been no studies on age-related
polymorphisms of HERVs in the human genome. HERV-K elements are the
youngest and most active family among HERVs. There has been
specific focus on the ability of these elements to cause or prevent
diseases through their expression (7). Taking these into consideration, the
present study detected retrotransposon polymorphisms in HERV-K
member 6 (HERV-K6) and HERV-K11 in healthy individuals of different
ages (between 10 and 79 years old) by using the
inter-retrotransposon amplified polymorphism (IRAP) molecular
marker technique. The IRAP technique was developed as a molecular
marker method due to the abundance and ubiquity of long terminal
repeat (LTR) transposons in the plant genome (8). This technique amplifies the sequences
between two adjacent retrotransposons with primers facing outward
from the LTR sequences (9). Previous
studies by our group demonstrated that the IRAP technique could
also be applied to the human genome (10,11). Thus,
the IRAP technique was the method of choice in the present study
for polymorphism analysis of HERV-K transpositions in human
subjects.
Materials and methods
Sample collection and DNA
extraction
HERV-K6 and HERV-K11 retrotransposon transpositions
were analysed in DNA samples of 18 healthy individuals between the
ages of 10 and 79 years old. The DNA samples were from Dr Kaniye
Sahin's DNA collections at Istanbul University Medical Faculty
(Istanbul, Turkey). The subjects from which samples were collected
were Turkish, and from the Istanbul University Department of
Molecular Biology and Genetics. Subjects completed an information
form prior to sample collection. This form requested health
information from subjects regarding concomitant diseases,
medication and genetic history. Excluded subjects were those
presenting with any disease or genetic disorder and/or were on a
current course of medication. Table I
lists the gender and age information of the subjects. The present
study established 5 age groups: 10–19 (n=4), 20–29 (n=4), 40–49
(n=3), 60–69 (n=3) and 70–79 (n=4) years. Genomic DNAs were
extracted from 5 ml venous blood samples (intravenous; collected
between January and February 2013) using a High Pure PCR Template
Preparation kit (Roche Diagnostics GmbH, Mannheim, Germany)
according to the manufacturer's protocol. The extracted genomic DNA
samples were stored at −20°C until use. The procedures followed
were in accordance with the current ethical standards of Istanbul
University Medical Faculty, and the information form included a
signed statement of written informed consent agreeing to the use of
patient materials for research purposes on the condition of
anonymity being retained.
 | Table I.Demographic information of enrolled
subjects. |
Table I.
Demographic information of enrolled
subjects.
| Subject no. | Sex | Age range
(years) |
|---|
| 1 | Female | 10–19 |
| 2 | Male | 10–19 |
| 3 | Female | 10–19 |
| 4 | Female | 10–19 |
| 5 | Male | 20–29 |
| 6 | Male | 20–29 |
| 7 | Male | 20–29 |
| 8 | Female | 20–29 |
| 9 | Male | 40–49 |
| 10 | Female | 40–49 |
| 11 | Male | 40–49 |
| 12 | Male | 60–69 |
| 13 | Female | 60–69 |
| 14 | Female | 60–69 |
| 15 | Female | 70–79 |
| 16 | Female | 70–79 |
| 17 | Female | 70–79 |
| 18 | Male | 70–79 |
IRAP-polymerase chain reaction (PCR)
analysis
Primer sequences of HERV-K6 and HERV-K11 were
obtained from the National Centre for Biotechnology Information
database (https://www.ncbi.nlm.nih.gov/; accession nos.
AF074086.2 and DQ112099.1, respectively; Table II). IRAP-PCR was performed with 2X
SapphireAmp Fast PCR Master Mix (Takara Biotechnology Co., Ltd.,
Dalina, China; RR350A), 10 µM of each primer and 20 ng template
genomic DNA. PCR amplification was performed under the following
cycling conditions: Denaturation at 95°C for 10 min, followed by 40
cycles of 94°C for 30 sec, 53–56°C for 30 sec and 72°C for 3 min,
and a final extension step at 72°C for 10 min (T100TM Thermal
Cycler; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The PCR
products were resolved by 2% agarose gel electrophoresis with
ethidium bromide staining. Following electrophoresis, the gel was
scanned and photographed on a UV transilluminator.
 | Table II.Primer sequences for HERV-K6 and
HERV-K11. |
Table II.
Primer sequences for HERV-K6 and
HERV-K11.
| HERV-K member | Primer | Sequence (5′-
3′) | Ta (°C) | Accession no. |
|---|
| HERV-K6 | Forward |
CCTACAGGTTTCACCATCTTG | 53 | AF074086.2 |
|
| Reverse |
CTTCTTTCTACACAGACACAG |
|
|
| HERV-K11 | Forward |
CCACAGGTGTGGAGGGACAACC | 56 | DQ112099.1 |
|
| Reverse |
CACCGAGACATTCCATTGCCC |
|
|
Determination of polymorphism
rates
The polymorphism ratios of samples were calculated
using the Jaccard similarity coefficient (12). In brief, bands were scored as a binary
value: ‘0’ for absence and ‘1’ for presence; the binary matrix
(1/0) was then used to calculate the similarity between the
different individuals using Jaccard's coefficient. Additionally,
the GelJ v.2.0 programme (Department of Mathematics and Computer
Science, University of La Rioja, Logroño, Spain) was used to
construct a phylogenetic tree: The unweighted pair group method
with arithmetic mean (UPGMA) clustering method of GelJ was used for
the gel images to construct dendrograms (13).
Results
Polymorphism analysis
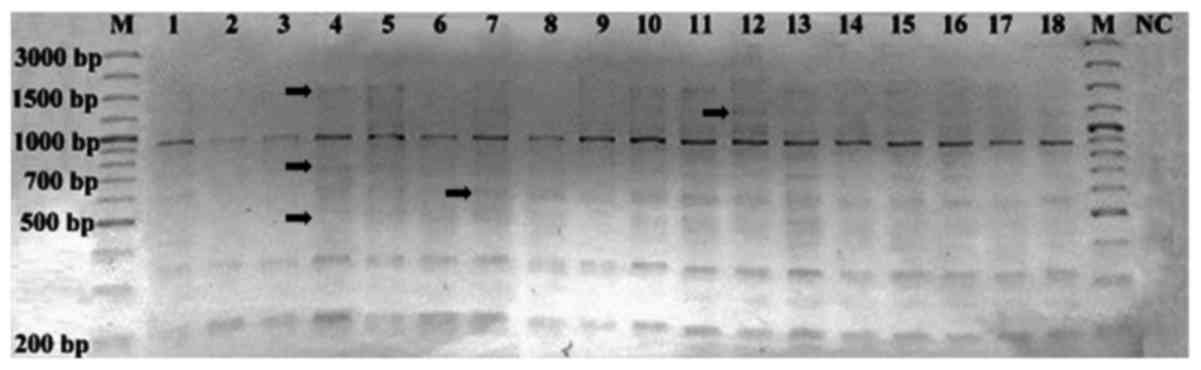
According to the band profiles of HERV-K6, a total
of 198 bands were detected, of which 137 were monomorphic bands and
61 were polymorphic bands, ranging from 200 to 3,000 bp (Fig. 1). The polymorphic (−) and monomorphic
(+) band numbers in each sample are listed in Table III. As a result of IRAP-PCR,
polymorphism ratios were determined as 0–70% for all samples
(Table IV). The polymorphism rates
were 0–70% among females and 11–60% among males. Age-associated
polymorphism was not observed in the study group (data not
shown).
 | Table III.Polymorphic (−) and monomorphic (+)
band numbers of HERV-K6. |
Table III.
Polymorphic (−) and monomorphic (+)
band numbers of HERV-K6.
|
| HERV-K6 |
|---|
|
|
|
|---|
| Subject no. | + | − |
|---|
| 1 | 5 | 6 |
| 2 | 4 | 7 |
| 3 | 4 | 7 |
| 4 | 8 | 3 |
| 5 | 9 | 2 |
| 6 | 7 | 4 |
| 7 | 9 | 2 |
| 8 | 6 | 5 |
| 9 | 6 | 5 |
| 10 | 9 | 2 |
| 11 | 10 | 1 |
| 12 | 10 | 1 |
| 13 | 10 | 1 |
| 14 | 8 | 3 |
| 15 | 8 | 3 |
| 16 | 8 | 3 |
| 17 | 8 | 3 |
| 18 | 8 | 3 |
 | Table IV.Polymorphism rates (%) of human
endogenous retrovirus type K member 6 determined by Jaccard
coefficient. |
Table IV.
Polymorphism rates (%) of human
endogenous retrovirus type K member 6 determined by Jaccard
coefficient.
| Subject no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|
| 1 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 2 | 20 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 | 20 | 0 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 4 | 56 | 50 | 50 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 5 | 60 | 56 | 56 | 11 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 6 | 50 | 43 | 43 | 13 | 22 | – |
|
|
|
|
|
|
|
|
|
|
|
|
| 7 | 44 | 56 | 56 | 30 | 36 | 22 | – |
|
|
|
|
|
|
|
|
|
|
|
| 8 | 63 | 57 | 57 | 25 | 33 | 14 | 33 | – |
|
|
|
|
|
|
|
|
|
|
| 9 | 57 | 50 | 50 | 56 | 44 | 50 | 60 | 43 | – |
|
|
|
|
|
|
|
|
|
| 10 | 60 | 70 | 70 | 30 | 20 | 40 | 36 | 33 | 44 | – |
|
|
|
|
|
|
|
|
| 11 | 50 | 60 | 60 | 20 | 10 | 30 | 27 | 40 | 50 | 10 | – |
|
|
|
|
|
|
|
| 12 | 50 | 60 | 60 | 36 | 27 | 30 | 10 | 40 | 50 | 27 | 18 | – |
|
|
|
|
|
|
| 13 | 50 | 60 | 60 | 20 | 10 | 30 | 27 | 40 | 50 | 10 | 0 | 18 | – |
|
|
|
|
|
| 14 | 38 | 50 | 50 | 22 | 30 | 13 | 11 | 25 | 56 | 30 | 20 | 20 | 20 | – |
|
|
|
|
| 15 | 38 | 50 | 50 | 22 | 30 | 13 | 11 | 25 | 56 | 30 | 20 | 20 | 20 | 0 | – |
|
|
|
| 16 | 38 | 50 | 50 | 22 | 30 | 13 | 11 | 25 | 56 | 30 | 20 | 20 | 20 | 0 | 0 | – |
|
|
| 17 | 38 | 50 | 50 | 22 | 30 | 13 | 11 | 25 | 56 | 30 | 20 | 20 | 20 | 0 | 0 | 0 | – |
|
| 18 | 38 | 50 | 50 | 22 | 30 | 13 | 11 | 25 | 56 | 30 | 20 | 20 | 20 | 0 | 0 | 0 | 0 | – |
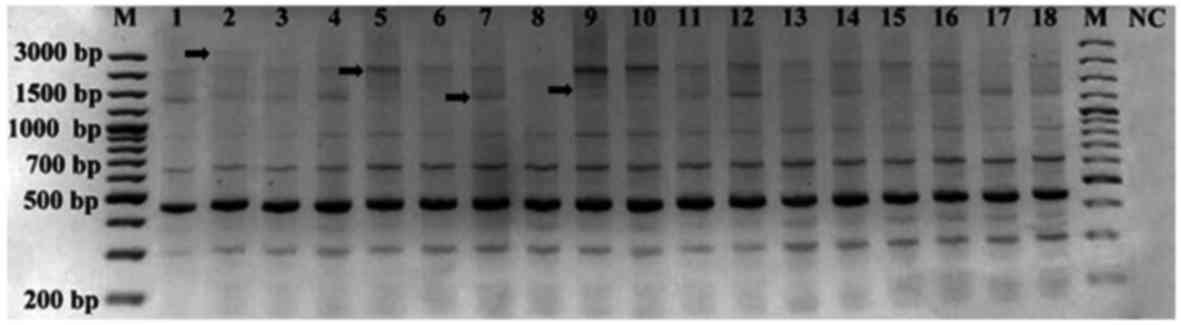
Analysis of HERV-K11 band profiles identified 162
scorable bands in all samples: 130 monomorphic bands and 32
polymorphic bands ranging from 200 to 3,000 bp (Fig. 2). The polymorphic (−) and monomorphic
(+) band numbers in each sample are listed in Table V. As a result of IRAP-PCR,
polymorphism ratios were determined as 0–38% for all samples
(Table VI). The polymorphism rates
were 0–38% among females and 0–25% among males. Similar to HERV-K6,
age-associated polymorphism was not observed in the study group
(data not shown).
 | Table V.Polymorphic (−) and monomorphic (+)
band numbers of HERV-K11. |
Table V.
Polymorphic (−) and monomorphic (+)
band numbers of HERV-K11.
|
| HERV-K11 |
|---|
|
|
|
|---|
| Subject no. | + | − |
|---|
| 1 | 7 | 2 |
| 2 | 8 | 1 |
| 3 | 7 | 2 |
| 4 | 7 | 2 |
| 5 | 8 | 1 |
| 6 | 6 | 3 |
| 7 | 7 | 2 |
| 8 | 5 | 4 |
| 9 | 8 | 1 |
| 10 | 8 | 1 |
| 11 | 7 | 2 |
| 12 | 8 | 1 |
| 13 | 8 | 1 |
| 14 | 7 | 2 |
| 15 | 7 | 2 |
| 16 | 8 | 1 |
| 17 | 7 | 2 |
| 18 | 7 | 2 |
 | Table VI.Polymorphism rates (%) of human
endogenous retrovirus type K member 11 determined by Jaccard
coefficient. |
Table VI.
Polymorphism rates (%) of human
endogenous retrovirus type K member 11 determined by Jaccard
coefficient.
| Subject no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|
| 1 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 2 | 13 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 | 0 | 13 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 4 | 0 | 13 | 0 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 5 | 13 | 22 | 13 | 13 | – |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 6 | 14 | 25 | 14 | 14 | 25 | – |
|
|
|
|
|
|
|
|
|
|
|
|
| 7 | 0 | 13 | 0 | 0 | 13 | 14 | – |
|
|
|
|
|
|
|
|
|
|
|
| 8 | 29 | 38 | 29 | 29 | 38 | 17 | 29 | – |
|
|
|
|
|
|
|
|
|
|
| 9 | 13 | 22 | 13 | 13 | 0 | 25 | 13 | 38 | – |
|
|
|
|
|
|
|
|
|
| 10 | 13 | 22 | 13 | 13 | 0 | 25 | 13 | 38 | 0 | – |
|
|
|
|
|
|
|
|
| 11 | 0 | 13 | 0 | 0 | 13 | 14 | 0 | 29 | 13 | 13 | – |
|
|
|
|
|
|
|
| 12 | 13 | 22 | 13 | 13 | 0 | 25 | 13 | 38 | 0 | 0 | 13 | – |
|
|
|
|
|
|
| 13 | 13 | 22 | 13 | 13 | 0 | 25 | 13 | 38 | 0 | 0 | 13 | 0 | – |
|
|
|
|
|
| 14 | 0 | 13 | 0 | 0 | 13 | 14 | 0 | 29 | 13 | 13 | 0 | 13 | 13 | – |
|
|
|
|
| 15 | 0 | 13 | 0 | 0 | 13 | 14 | 0 | 29 | 13 | 13 | 0 | 13 | 13 | 0 | – |
|
|
|
| 16 | 13 | 22 | 13 | 13 | 0 | 25 | 13 | 38 | 0 | 0 | 13 | 0 | 0 | 13 | 13 | – |
|
|
| 17 | 0 | 13 | 0 | 0 | 13 | 14 | 0 | 29 | 13 | 13 | 0 | 13 | 13 | 0 | 0 | 13 | – |
|
| 18 | 0 | 13 | 0 | 0 | 13 | 14 | 0 | 29 | 13 | 13 | 0 | 13 | 13 | 0 | 0 | 13 | 0 | – |
Clustering analysis
The UPGMA clustering method was performed for
HERV-K6 and HERV-K11 profiles in the samples. According to the band
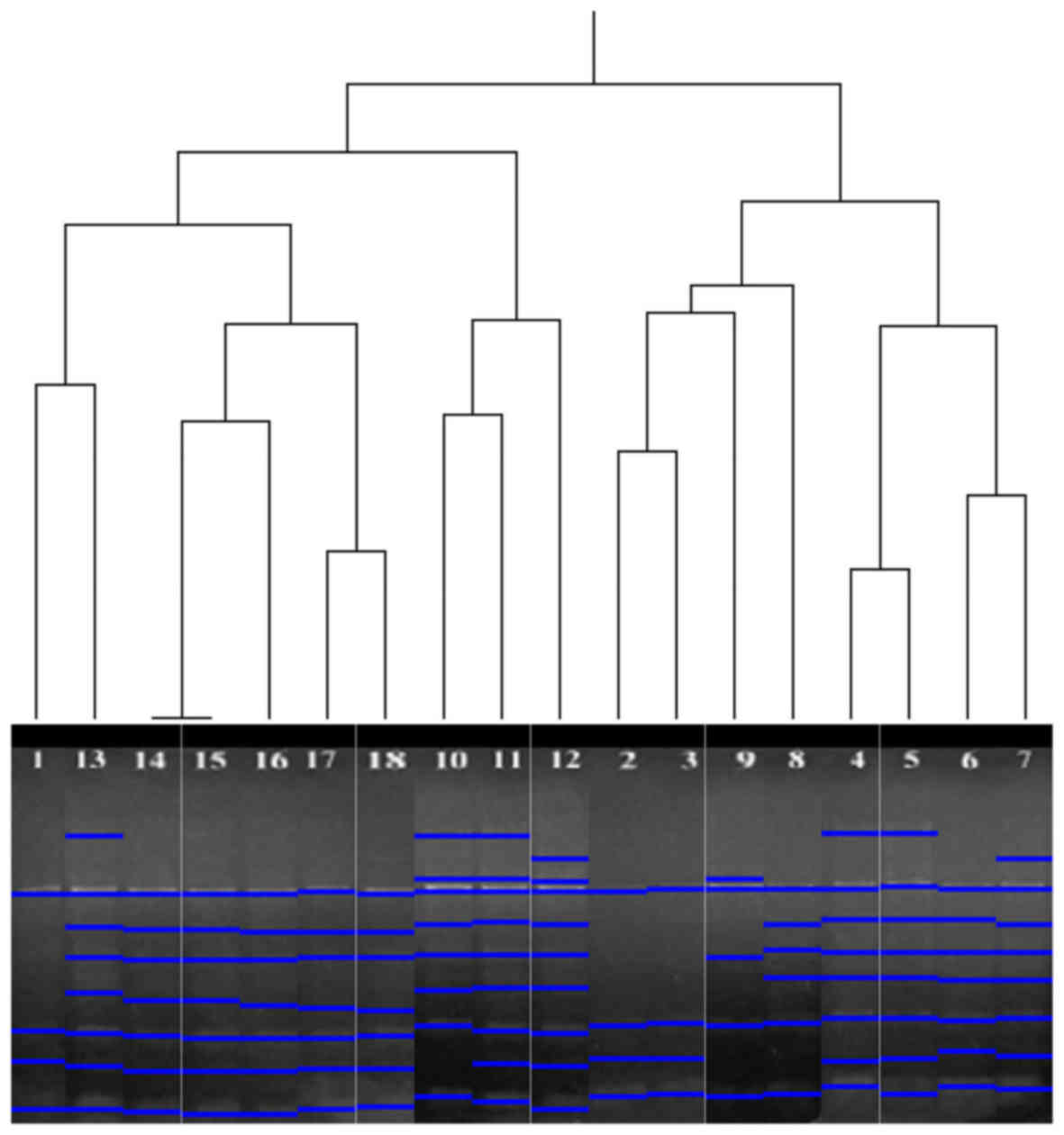
profiles of HERV-K6, the 18 analysed samples were grouped in two
clusters. The first group consisted of the numbered samples 1 and
10–18, while the second group consisted of the numbered samples 2–9
(Fig. 3). As a result of UPGMA
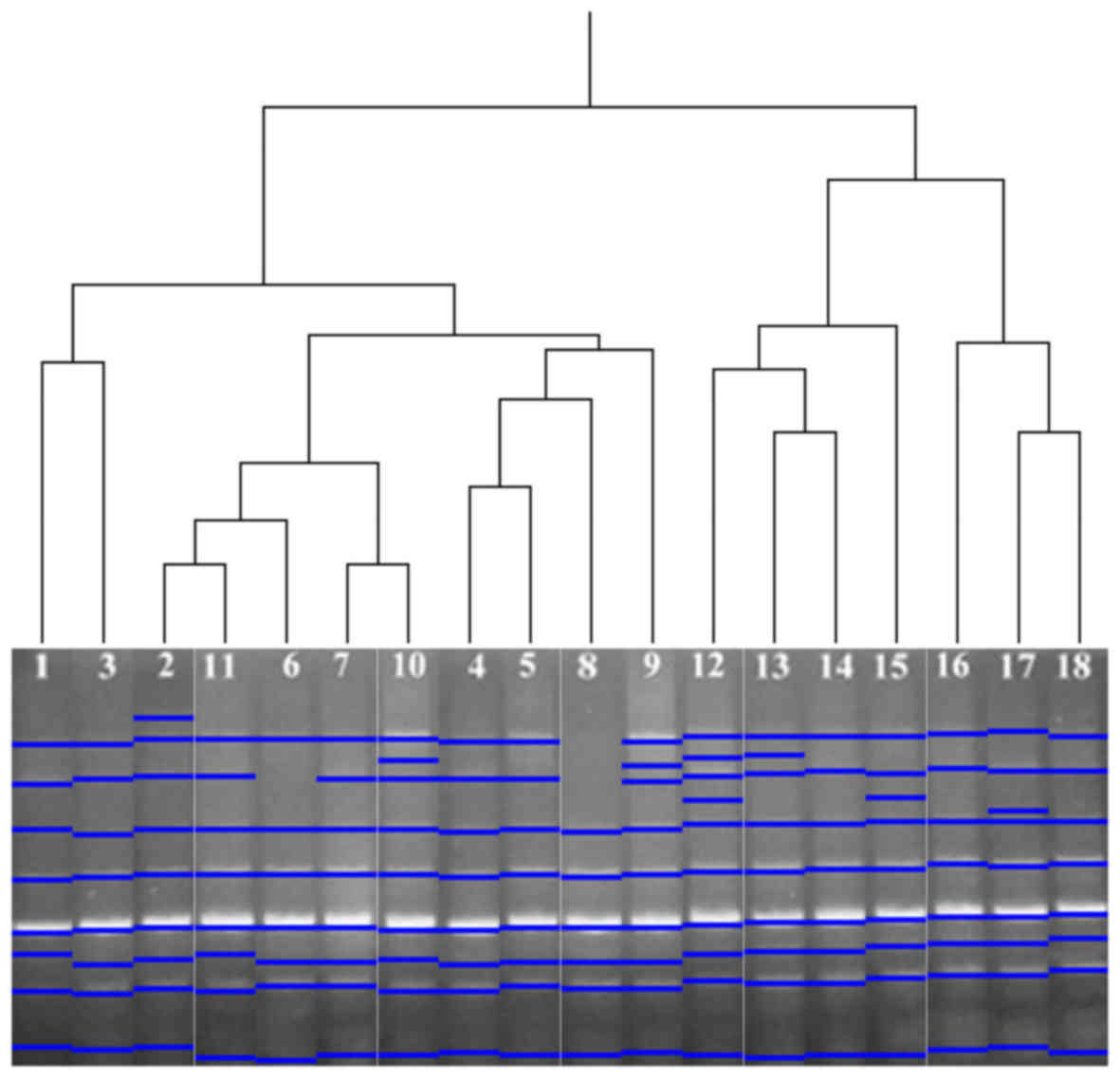
analysis of HERV-K11, the 18 analysed samples were grouped in two
clusters. Samples 1–11 were grouped in the first cluster, while
12–18 were grouped in the second (Fig.
4).
Discussion
The presence of HERVs in the human genome is
considered to be a result of insertion of their exogenous ancestors
into primate germ-line cells (14).
Further amplification via retrotranspositions likely resulted in
the formation of the HERV families identified to date, which have
been suggested to serve a significant role in primate evolution
(14–16). The HERV-K family is established as the
most functionally active group of endogenous retroviruses (17). In the present study, the IRAP
molecular marker technique was used to assess HERV-K6 and HERV-K11
transpositions in the human genome, and determined that HERV-K6 and
HERV-K11 elements may still be active in the human genome.
According to the IRAP analysis results, HERV-K6 and HERV-K11
exhibited notably similar band profiles (137 monomorphic bands for
HERV-K6; 130 monomorphic bands for HERV-K11). However, the small
numbers of polymorphic bands varied between the HERV-K6 and
HERV-K11 profiles of the different DNA samples (61 polymorphic
bands for HERV-K6; 32 polymorphic bands for HERV-K11). When band
profiles of samples were compared between different age groups and
same age groups, no marked differences were determined regarding
polymorphism age specificity. Furthermore, as the study group was
composed of males and females, the band patterns of males and
females were compared; however, no gender specific polymorphisms
were determined. Therefore, polymorphisms were considered to be
individual specific. Additionally, the study group did not include
any family members. Thus, polymorphism rates may also be family
specific.
There have been a number of studies on HERV
polymorphisms (11,18–20). A
study by Mamedov et al (18)
identified a novel HERV-K solo LTR insertion polymorphism,
suggesting a recent retrovirus insertion followed by a
recombination event between two retroviral LTRs. Additionally, the
allele frequencies were studied in 88 human DNA samples among
different ethnic groups: Mordvinians, Bashkirs and Kalmyks (all
from Russia) and five samples of African origin (all from
Guinea-Bissau) (18). Another study
by Guliyev et al (11)
observed integration polymorphism patterns for HERV-H in the tested
individuals (n=20) of diverse ethnic origin. Furthermore, a study
by Kahyo et al (19)
investigated HML-2 (HERV-K) insertional polymorphisms in a small
Japanese population. They compared reference genomes obtained from
genome projects and genomic PCR sequences. Sequencing of the
preintegration sites identified a HML-2 site, located at 7p21.2.
Another insertionally polymorphic site for a non-human-specific
HML-2 site was also identified at 6p25.2 (19). In a study conducted by Belshaw et
al (20), 113 human-specific
HERV-K (HML2) elements were identified in the human genome
sequence, 8 of which were insertionally polymorphic. Furthermore,
it has been determined that the number of polymorphic elements was
not significantly different from that predicted by a standard
population genetic model that assumes constant genetic activity of
the family (19). This suggests that
the HERV-K family may be active in humans.
According to the results of the present UPGMA
analysis, re-ordered samples exhibited similar band patterns. When
investigating the dendrograms of HERV-K6 and HERV-K11, it was
observed that all re-ordered samples were not included in the same
or different age groups (data not shown). For instance, samples 1,
13 and 14 were in the same group on UPGMA analysis of HERV-K6
profiles, despite the samples belonging to different age groups.
Similarly, while samples 6, 7 and 11 were in the same group on
UPGMA analysis of HERV-K11, they belonged to different age groups;
by contrast, samples 1–3 were in the same age group and also the
same group determined by UPGMA clustering. These results indicated
that the polymorphisms were not age-associated. Thus, the
polymorphisms may be individual specific.
High-throughput sequencing technologies have also
provided information on insertionally polymorphic sites of HERV-K
in different studies (21–23). A study by Shin et al (24) analysed insertions of HERV-K elements
including HERV-K101 and -K132. They concluded that HERV-K activity
served an important role in genomic divergence within the human
population. Movements of human retrotransposons may differ between
normal somatic tissue and somatic tumour genomes (25). The present HERV-K6 and HERV-K11
insertion polymorphism analyses were similar to those reported
previously with regard to determining insertion polymorphisms,
while they differ from some studies due to the different techniques
used (11,18–21).
Guliyev et al (11) determined
polymorphisms with the IRAP technique whereas Mamedov et al
(18) tested distribution of the
LTR-containing allele in Africans and Russian populations. Kahyo
et al (19) identified
insertional polymorphisms with genomic PCR analysis in a Japanese
group. Belshaw et al (20) and
Lee et al (21) determined
insertional polymorphisms with sequencing and bioinformatics.
HERVs are inactivated by mutations, deletions or
recombinations (26). Studies have
indicated that certain active copies may express proteins or
virions, and have pathogenic effects or physiological roles
(2). Furthermore, there may be an
association between the expression of HERV-K elements and the
development of cancer and autoimmune diseases (24). A study performed by Li et al
(27) detected HERV-K envelope
protein expression in pancreatic cancer cell lines and patient
biopsies, but not in normal pancreatic cell lines or uninvolved
normal tissues. Maze et al (28) demonstrated that HERV-K envelope,
capsid, Rec and Np9 proteins were overexpressed in human primary
schwannoma cells and tissues. They also identified that anti-HERV-K
antibodies reduced p53 expression and schwannoma proliferation;
furthermore, pre-incubation of schwannoma cells with HERV-K
antibodies prior to treatment with cancer drugs (AZD6244
with/without Sorafenib and BEZ235) potentiated the drug efficiency.
Thus, they suggested that HERV-K has a pathogenic role in
schwannoma and may be a promising therapeutic target (28). Additionally, a HERV-K-related insert
may serve as an enhancer for the schizophrenia-linked gene proline
dehydrogenase, a candidate gene for sensitivity to schizophrenia
and other neurological diseases (29). Another clinical study on HERV-K
indicated that HERV-K expression was markedly higher in leukaemia
patients when compared with its expression in healthy donors of a
similar median age (30). To date,
HERV studies have analysed the association between expression and
neurological diseases, cancer or autoimmune diseases (24). There have been few reports related to
HERV retrotransposon movements in the human genome (31). To the best of our knowledge, the
present analyses were the first to focus on HERV-K6 and HERV-K11
transpositions in different healthy individuals. In the present
study, polymorphisms were also investigated in healthy subjects
according to different age groups. Although polymorphism was
identified among all subjects, the HERV-K6 and HERV-K11 endogenous
retrovirus polymorphisms exhibited no age-associations.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Istanbul
University Scientific Research Projects Coordination Unit (grant
no. 22142).
Availability of data and materials
The DNA samples were from Dr Kaniye Sahin's DNA
collections at Istanbul University Medical Faculty (Istanbul,
Turkey). All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BCG, EK and SM analysed and interpreted the data,
and wrote the draft manuscript. NG revised the manuscript for
important intellectual content and gave final approval of the
version to be published. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Patient donors of the DNA samples provided written
informed consent agreeing to the use of patient materials for
research purposes.
Consent for publication
Patient donors of the DNA samples provided written
informed consent permitting publication of relevant data on the
condition of anonymity being retained.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HERVs
|
human endogenous retroviruses
|
|
HERV-K
|
human endogenous retrovirus type K
|
|
HML-2
|
human mouse mammary tumour virus
like-2
|
|
IRAP
|
inter-retrotransposon amplified
polymorphism
|
|
LTR
|
long terminal repeat
|
|
PCR
|
polymerase chain reaction
|
|
UPGMA
|
unweighted pair group method with
arithmetic mean
|
References
|
1
|
Perron H and Lang A: The human endogenous
retrovirus link between genes and environment in multiple sclerosis
and in multifactorial diseases associating neuroinflammation. Clin
Rev Allergy Immunol. 39:51–61. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Subramanian RP, Wildschutte JH, Russo C
and Coffin JM: Identification, characterization, and comparative
genomic distribution of the HERV-K (HML-2) group of human
endogenous retroviruses. Retrovirology. 8:902011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Downey RF, Sullivan FJ, Wang-Johanning F,
Ambs S, Giles FJ and Glynn SA: Human endogenous retrovirus K and
cancer: Innocent bystander or tumorigenic accomplice? Int J Cancer.
137:1249–1257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonzalez-Cao M, Iduma P, Karachaliou N,
Santarpia M, Blanco J and Rosell R: Human endogenous retroviruses
and cancer. Cancer Biol Med. 13:483–488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hurst TP and Magiorkinis G: Epigenetic
control of human endogenous retrovirus Expression: Focus on
regulation of long-terminal repeats (LTRs). Viruses. 9:92017.
View Article : Google Scholar
|
|
6
|
Kremer D, Glanzman R, Traboulsee A, Nath
A, Groc L, Horwitz M, Göttle P, Perron H, Gold J, Hartung HP, et
al: Prehistoric enemies within: The contribution of human
endogenous retroviruses to neurological diseases. Meeting report:
‘Second International Workshop on Human Endogenous Retroviruses and
Disease’, Washington DC, March 13th and 14th 2017. Mult Scler Relat
Disord. 15:18–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Griffiths DJ: Endogenous retroviruses in
the human genome sequence. Genome Biol. 2:S10172001. View Article : Google Scholar
|
|
8
|
Kalendar R, Grob T, Regina M, Suoniemi A
and Schulman A: IRAP and REMAP: Two new retrotransposon-based DNA
fingerprinting techniques. Theor Appl Genet. 98:704–711. 1999.
View Article : Google Scholar
|
|
9
|
Gozukirmizi N, Yilmaz S, Marakli S and
Temel A: Retrotransposon-based molecular markers; Tools for
variation analysis in plantsApplications of Molecular Markers in
Plant Genome analysis and Breeding. Taski-Ajdukovic K: Research
Signpost; Kerala: pp. 19–45. 2015
|
|
10
|
Cakmak B, Marakli S and Gözükirmizi N:
Sukkula retrotransposon movements in the human genome. Biotechnol
Biotechnol Equip. 31:900–905. 2017.
|
|
11
|
Guliyev M, Yılmaz S, Şahin K, Maraklı S
and Gözükirmizi N: Human endogenous retrovirus-H insertion
screening. Mol Med Rep. 7:1305–1309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jaccard P: Nouvelles recherches sur la
distribution florale. Bull Soc Vaud Sci Nat. 44:223–270. 1908.
|
|
13
|
Heras J, Domínguez C, Mata E, Pascual V,
Lozano C, Torres C and Zarazaga M: GelJ - a tool for analyzing DNA
fingerprint gel images. BMC Bioinformatics. 16:2702015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sverdlov ED: Retroviruses and primate
evolution. BioEssays. 22:161–171. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van de Lagemaat LN, Landry JR, Mager DL
and Medstrand P: Transposable elements in mammals promote
regulatory variation and diversification of genes with specialized
functions. Trends Genet. 19:530–536. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kazazian HH Jr: Mobile elements: Drivers
of genome evolution. Science. 303:1626–1632. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seifarth W, Spiess B, Zeilfelder U, Speth
C, Hehlmann R and Leib-Mösch C: Assessment of retroviral activity
using a universal retrovirus chip. J Virol Methods. 112:79–91.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mamedov I, Lebedev Y, Hunsmann G,
Khusnutdinova E and Sverdlov E: A rare event of insertion
polymorphism of a HERV-K LTR in the human genome. Genomics.
84:596–599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kahyo T, Yamada H, Tao H, Kurabe N and
Sugimura H: Insertionally polymorphic sites of human endogenous
retrovirus-K (HML-2) with long target site duplications. BMC
Genomics. 18:4872017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Belshaw R, Dawson ALA, Woolven-Allen J,
Redding J, Burt A and Tristem M: Genomewide screening reveals high
levels of insertional polymorphism in the human endogenous
retrovirus family HERV-K(HML2): Implications for present-day
activity. J Virol. 79:12507–12514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee E, Iskow R, Yang L, Gokcumen O,
Haseley P, Luquette LJ III, Lohr JG, Harris CC, Ding L, Wilson RK,
et al: Cancer Genome Atlas Research Network: Landscape of somatic
retrotransposition in human cancers. Science. 337:967–971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marchi E, Kanapin A, Magiorkinis G and
Belshaw R: Unfixed endogenous retroviral insertions in the human
population. J Virol. 88:9529–9537. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wildschutte JH, Williams ZH, Montesion M,
Subramanian RP, Kidd JM and Coffin JM: Discovery of unfixed
endogenous retrovirus insertions in diverse human populations. Proc
Natl Acad Sci USA. 113:E2326–E2334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shin W, Lee J, Son SY, Ahn K, Kim HS and
Han K: Human-specific HERV-K insertion causes genomic variations in
the human genome. PLoS One. 8:e606052013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kano H, Godoy I, Courtney C, Vetter MR,
Gerton GL, Ostertag EM and Kazazian HH Jr: L1 retrotransposition
occurs mainly in embryogenesis and creates somatic mosaicism. Genes
Dev. 23:1303–1312. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vogel G: The human genome. Objection #2:
Why sequence the junk? Science. 291:11842001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Radvanyi L, Yin B, Li J, Chivukula
R, Lin K, Lu Y, Shen J, Chang DZ, Li D, et al: Down-regulation of
human endogenous retrovirus type K (HERV-K) viral env RNA in
pancreatic cancer cells decreases cell proliferation and tumour
growth. Clin Cancer Res. 23:5892–5911. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maze E, Reeves S, Hilton D, Provenzano L,
Belshaw R and Ammoun S: Abstract 4627: The role of human endogenous
retroviral proteins in the development of Merlin-deficient tumors
and as potential drug targets. In: Proceedings of the AACR 107th
Annual Meeting 2016, New Orleans, LA. Cancer Res. 76 Suppl
14:Abstract nr 4627. 2016.
|
|
29
|
Suntsova M, Gogvadze EV, Salozhin S,
Gaifullin N, Eroshkin F, Dmitriev SE, Martynova N, Kulikov K,
Malakhova G, Tukhbatova G, et al: Human-specific endogenous
retroviral insert serves as an enhancer for the
schizophrenia-linked gene PRODH. Proc Natl Acad Sci USA.
110:19472–19477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bergallo M, Montanari P, Mareschi K,
Merlino C, Berger M, Bini I, Daprà V, Galliano I and Fagioli F:
Expression of the pol gene of human endogenous retroviruses HERV-K
and -W in leukemia patients. Arch Virol. 162:3639–3644. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iskow RC, McCabe MT, Mills RE, Torene S,
Pittard WS, Neuwald AF, Van Meir EG, Vertino PM and Devine SE:
Natural mutagenesis of human genomes by endogenous
retrotransposons. Cell. 141:1253–1261. 2010. View Article : Google Scholar : PubMed/NCBI
|