Introduction
Diabetic peripheral neuropathy (DPN) is a common
complication of diabetes mellitus (DM). More than half of patients
with DM worldwide suffer from DPN during the course of the disease
(1,2).
DPN is characterized by distal-to-proximal nerve damage leading to
neuropathic pain and loss of sensation (3). It is also associated with notably high
morbidity and mortality: Previous studies reported that the
mortality rate approximated to 25–50% within 5–10 years following
onset of diabetic neuropathy (4,5). Strict
maintenance of normal glycemic level is the only effective
treatment available for DPN at present (6).
Though the occurrence of DPN is common to both type
1 (T1) and type 2 (T2)DM, a number of studies have suggested that
its pathogenic mechanism may differ between the two (7,8). For
example, more abnormalities at the molecular, functional and
morphometric levels including increased frequencies of denervated
Schwann cells and significant fiber loss have been observed in the
peripheral nerve of T1DM mice compared with in T2DM mice (8,9). In
addition, different structural changes may lead to variation in
nerve conduction velocity for DPN in the two diabetic models
(10). These fundamentally different
mechanisms for DPN may lead to varying effects of the common
treatments, such as in control of glucose, which has been suggested
to be more beneficial for DPN in T1DM than in T2DM (7). Therefore, determining the distinct
molecular mechanisms underlying DPN in T1- and T2DM is of paramount
importance for the development of successful therapeutic
interventions.
In the last decade, the development of
high-throughput platforms including microarray technology has
allowed researchers to concurrently determine the expression levels
of several thousands of differentially expressed genes (DEGs) in
diseases (11,12). In addition, the comparison of gene
expression profiles of DEGs through microarray technology using
bioinformatics analysis has determined distinct pathophysiological
mechanisms in different diseases or syndromes including neuropathic
pain and chronic radicular pain (13). For instance, Zhang et al
(14) screened a number of DEGs in a
control (healthy) group of mice and a group with streptozotocin
(STZ)-induced diabetes, and identified genes co-regulated by both
STZ and rosiglitazone, which may be potential targets in the
treatment of DPN. However, there are few studies that have compared
the gene expression profiles of DEGs between DPN in T1- and T2DM
through microarray profiling. In the present study, the aim was to
compare the DEGs between the sciatic nerves of T1- and T2DM mouse
models by microarray profiling. Furthermore, the distinct
biological processes and pathways associated with DPN in T1- or
T2DM were analyzed and compared based on gene ontology (GO) and
pathway enrichment analyses. This was hoped to provide novel
insights into the distinct pathophysiological mechanisms and
implicate drug therapies for DPN specific to the different types of
DM.
Materials and methods
Source of microarray data
The gene expression profiles of GSE11343 and
GSE27382 were obtained from the Gene Expression Omnibus (GEO)
database (http://www.ncbi.nlm.nih.gov/geo/). The annotation
platforms for GSE11343 and GSE27382 were GPL1261 and GPL9746,
respectively.
For GSE11343 submitted by Wiggin et al
(15), included datasets were of five
sciatic nerve samples from mice with T1DM induced by STZ
(GSM286169, GSM286173, GSM286176, GSM286178 and GSM286430) and four
sciatic nerve samples from normal mice (GSM286159, GSM286160,
GSM286163 and GSM286165).
For GSE27382 submitted by Pande et al
(16), included datasets were of six
sciatic nerve samples from db/db mice with T2DM
(GSM677112-GSM677117) and seven sciatic nerve samples from db/+
(normal) mice (GSM677105-GSM677111).
Pre-processing of microarray data and
identification of DEGs
Pre-processing for the cell intensity (CEL) files
including conversion into expression measures, background
correction and quartile data normalization was performed with
BRB-ArrayTools (version 4.5.1) (17).
The univariate t-test with a fold change ≥2 and nominal
significance level of 0.05 was applied in BRB-ArrayTools to
identify the DEGs between the diabetic and normal groups.
Gene ontology and pathway enrichment
analyses
To identify the DEGs determined with BRB-ArrayTools,
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyses were performed via the Database for Annotation,
Visualization and Integrated Discovery (DAVID 6.8; http://david.abcc.ncifcrf.gov/) (18,19). GO
terms [categorized into molecular function (MF), biological process
(BP) and cellular component (CC)] and KEGG pathways with P<0.05
were considered significantly enriched by the DEGs.
Construction of protein-protein
interaction (PPI) network and module analysis
With the purpose of evaluating the relationships
among DEGs from the perspective of protein interaction, a
protein-protein interaction (PPI) network was constructed with the
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING
10.5; http://www.string-db.org) and visualized
using Cytoscape software (version 3.4.0; http://cytoscape.org/) (18). A combined score >0.4 was selected
to determine significant interactions among DEGs. In accordance
with a previous analysis (20), the
connectivity degree of a protein, namely the number of proteins it
connected with, was considered to indicate importance of the
protein in the PPI network.
Module analysis was performed in the plugin
ClusterONE (version 1.0) in Cytoscape with a threshold of
P<0.001. In addition, function and pathway enrichment analyses
were performed for DEGs in the modules with a threshold of
P<0.05.
Results
Identification of DEGs
Based on the criteria of a nominal significance
level of 0.05 and fold change ≥2, 623 and 1,890 DEGs were
identified in sciatic nerves of T1DM (GSE11343) and T2DM (GSE27382)
mice, respectively, as compared with the control samples. Among
them, 75 genes were identified to be coordinately dysregulated in
sciatic nerves of both models, with 20 genes upregulated and 55
genes downregulated (Fig. 1 and
Table SI). Meanwhile, 160
upregulated and 388 downregulated DEGs were unique to T1DM
(Fig. 1 and Table SII), and 721 upregulated and 1,094
downregulated DEGs were unique to T2DM (Fig. 1 and Table
SIII).
 | Table SI.List of genes coordinately regulated
in type 1 and type 2 diabetes mellitus. |
Table SI.
List of genes coordinately regulated
in type 1 and type 2 diabetes mellitus.
| Gene count | Gene symbol | Gene count | Gene symbol | Gene count | Gene symbol |
|---|
| Upregulated |
| 1 | C1qtnf9 | 8 | Myo5b | 15 | Hipk3 |
| 2 | Hspb7 | 9 | Tcf15 | 16 | Hif3a |
| 3 | Fmo2 | 10 | Ltbp2 | 17 | Pde2a |
| 4 | Cyp1a1 | 11 | Tlr7 | 18 | Fkbp5 |
| 5 | Gpihbp1 | 12 | Map3k6 | 19 | 8430436 N08Rik |
| 6 | Mbd1 | 13 | Angptl4 | 20 | Dnah11 |
| 7 | Gm4876 | 14 | D8Ertd82e |
|
|
| Downregulated |
| 1 | Kcna2 | 20 | Pcyox1l | 39 | Tmem229a |
| 2 | Kif5b | 21 | Rapgef4 | 40 | Rian |
| 3 | Cct4 | 22 | Sptssb | 41 | C1ql3 |
| 4 | Fchsd2 | 23 | 5930427L02Rik | 42 | Chic1 |
| 5 | Fgf14 | 24 | Efcab14 | 43 | Cntn3 |
| 6 | Fndc5 | 25 | Frzb | 44 | Map1b |
| 7 | Kcna1 | 26 | Cemip | 45 | Slc35f1 |
| 8 | Slc25a27 | 27 | Zic2 | 46 | Sox9 |
| 9 | Galnt3 | 28 | Zic1 | 47 | Gpm6b |
| 10 | Ncald | 29 | Slc1a3 | 48 | Slc47a1 |
| 11 | Senp8 | 30 | Snhg14 | 49 | Elavl2 |
| 12 | Asph | 31 | Bmp5 | 50 | Ptn |
| 13 | Pdia3 | 32 | Lamc3 | 51 | Dcc |
| 14 | Bicd1 | 33 | Nipal1 | 52 | Atp13a5 |
| 15 | Elovl4 | 34 | C78859 | 53 | Hpca |
| 16 | Npas3 | 35 | Glipr2 | 54 | Sox8 |
| 17 | Opcml | 36 | Enah | 55 | Zic5 |
| 18 | Nr2f2 | 37 | Gm19935 |
|
|
| 19 | Ntrk3 | 38 | Paqr8 |
|
|
 | Table SII.List of genes specific to type 1
diabetes mellitus. |
Table SII.
List of genes specific to type 1
diabetes mellitus.
| Gene count | Gene symbol | Gene count | Gene symbol | Gene count | Gene symbol |
|---|
| Upregulated |
|
1 | Myh2 | 55 | Wdr63 | 108 | Bbd9 |
|
2 | Doc2b | 56 | Gm17455 | 109 | Klk1b24 |
|
3 | Atxn7l1 | 57 | Per2 | 110 | Acer2 |
|
4 | Tbata | 58 | Gimap8 | 111 | Angpt2 |
|
5 | Rph3al | 59 | Cpa3 | 112 | BC003965 |
|
6 | Apoc3 | 60 | C77370 | 113 | Rtp3 |
|
7 | Nr5a2 | 61 | Rgs3 | 114 | Iffo1 |
|
8 | Alpk3 | 62 | Oosp1 | 115 | Lhx6 |
|
9 | Tal1 | 63 | Dbp | 116 | Tmem88 |
| 10 | Aldh3b2 | 64 | Dll4 | 117 | 4930539E08Rik |
| 11 | Rasd2 | 65 | Mapk12 | 118 | Adcyap1r1 |
| 12 | Iigp1 | 66 | D7Bwg0826e | 119 | Anapc11 |
| 13 | Etnppl | 67 | Myct1 | 120 | Clca3a2 |
| 14 | Colec11 | 68 | Ranbp17 | 121 | Cyyr1 |
| 15 | Ifi44 | 69 | Adcy4 | 122 | Lrpap1 |
| 16 | Krt18 | 70 | Shisa6 | 123 | Cdh13 |
| 17 | Gimap4 | 71 | Asxl1 | 124 | Abcg1 |
| 18 | Adgrf5 | 72 | Ppp1r3d | 125 | Syt7 |
| 19 | Spns2 | 73 | Camk2n1 | 126 | Jag2 |
| 20 | Cabp4 | 74 | Mboat1 | 127 | Odf3b |
| 21 | D7Wsu130e | 75 | Arrdc4 | 128 | Rilp |
| 22 | Ms4a2 | 76 | Ctsg | 129 | Sox7 |
| 23 | Exoc3l2 | 77 | Pvr | 130 | 9430030N17Rik |
| 24 | Ms4a8a | 78 | Tox | 131 | Adgrl4 |
| 25 | Csf2rb2 | 79 | 4833438C02Rik | 132 | Usp3 |
| 26 | Ace2 | 80 | Rasgrf2 | 133 | Vps51 |
| 27 | Itih4 | 81 | Tnfsf13os | 134 | Kcnq1 |
| 28 | Robo4 | 82 | Gm11772 | 135 | Krt20 |
| 29 | Gm11716 | 83 | 4833411I10Rik | 136 | Rabgap1l |
| 30 | 4921507P07Rik | 84 | Mfng | 137 | Sphk2 |
| 31 | Gp1bb | 85 | Sox17 | 138 | 1810021B22Rik |
| 32 | Vwf | 86 | D930003E18Rik | 139 | 4930447M23Rik |
| 33 | Erg | 87 | Rtn4rl2 | 140 | Arhgef15 |
| 34 | Eng | 88 | Kank3 | 141 | Cpsf1 |
| 35 | Mettl22 | 89 | Ptprb | 142 | Foxp4 |
| 36 | Lama3 | 90 | Rpusd2 | 143 | Sema5b |
| 37 | 5730458M16Rik | 91 | Sigirr | 144 | Actr1b |
| 38 | 4833403J16Rik | 92 | Trim47 | 145 | Hsd17b11 |
| 39 | Tceanc2 | 93 | Adamdec1 | 146 | Myrip |
| 40 | Fam107a | 94 | Cgrrf1 | 147 | Gimap5 |
| 41 | Slc27a2 | 95 | Tigd3 | 148 | Prkcg |
| 42 | Zfpm1 | 96 | Spn | 149 | Abi3 |
| 43 | Clec14a | 97 | Car7 | 150 | Bmp4 |
| 44 | Oasl1 | 98 | Mettl13 | 151 | Mgst2 |
| 45 | Anxa8 | 99 | Rap1gap | 152 | Ppp1r13l |
| 46 | D130037M23Rik | 100 | 2210019G11Rik | 153 | Selo |
| 47 | Fam189a1 | 101 | 6330403A02Rik | 154 | Sema3f |
| 48 | Clca3a1 | 102 | Tmprss2 | 155 | Mocs1 |
| 49 | Mmrn2 | 103 | Ak8 | 156 | Nos3 |
| 50 | Cbfa2t3 | 104 | Cyp11a1 | 157 | Pold1 |
| 51 | Notch4 | 105 | Ushbp1 | 158 | AU022252 |
| 52 | She | 106 | Slc6a7 | 159 | D630045J12Rik |
| 53 | Soat2 | 107 | 6330416G13Rik | 160 | Ngef |
| 54 | Unc45bos |
|
|
|
|
| Downregulated |
|
1 | Ccnt2 | 131 | 9530029O12Rik | 260 | Dag1 |
|
2 | Col4a5 | 132 | A330075M08Rik | 261 | Itm2a |
|
3 | Crkl | 133 | Abce1 | 262 | Luc7l2 |
|
4 | Dnajc18 | 134 | Anp32a | 263 | Marcks |
|
5 | Ebf2 | 135 | Arhgef6 | 264 | Nktr |
|
6 | Lamb1 | 136 | Chl1 | 265 | Saa1 |
|
7 | Pbrm1 | 137 | Cp | 266 | Senp6 |
|
8 | R3hdm4 | 138 | Ctse | 267 | Serinc1 |
|
9 | Rab10 | 139 | Dcbld2 | 268 | Sfrp1 |
| 10 | Rbfox2 | 140 | Eif4a1 | 269 | Slc25a24 |
| 11 | Rsl24d1 | 141 | Epb41l2 | 270 | Snx18 |
| 12 | Sat1 | 142 | Far1 | 271 | Snx5 |
| 13 | Slc25a3 | 143 | Gatm | 272 | Tbl1×r1 |
| 14 | Smarcc1 | 144 | Gpbp1l1 | 273 | Zic4 |
| 15 | Spry2 | 145 | Hba-a2 | 274 | 9930031P18Rik |
| 16 | Suz12 | 146 | Hmgcr | 275 | Akap13 |
| 17 | Timp2 | 147 | Igf1r | 276 | Dtx3l |
| 18 | Zfp626 | 148 | Klhl28 | 277 | Hsph1 |
| 19 | Actb | 149 | Mboat7 | 278 | Lin7c |
| 20 | Amd1 | 150 | Pddc1 | 279 | Peg3 |
| 21 | Arf1 | 151 | Ppp1r10 | 280 | Ppp4c |
| 22 | Arhgap29 | 152 | Prpf39 | 281 | Pum2 |
| 23 | Atp6v1b2 | 153 | Pura | 282 | Rad51c |
| 24 | C030046G05 | 154 | Rbm25 | 283 | Scd2 |
| 25 | Cbx5 | 155 | Rnd3 | 284 | Slc12a2 |
| 26 | Cd47 | 156 | Slc7a11 | 285 | Sp7 |
| 27 | Cryzl1 | 157 | Trps1 | 286 | Stk25 |
| 28 | D2Ertd640e | 158 | Ttc14 | 287 | Txnip |
| 29 | Elavl1 | 159 | Vma21 | 288 | Usp48 |
| 30 | Hist1h1c | 160 | Vps13a | 289 | Zfp825 |
| 31 | Il6st | 161 | Yipf4 | 290 | AA409587 |
| 32 | Katnbl1 | 162 | Add3 | 291 | B4galt6 |
| 33 | Lats2 | 163 | Atxn7l3b | 292 | D9Ertd292e |
| 34 | P4ha1 | 164 | BC052688 | 293 | Dusp3 |
| 35 | Pi4kb | 165 | C230076A16Rik | 294 | Klhl24 |
| 36 | Plekhg1 | 166 | Col8a1 | 295 | Msn |
| 37 | Polb | 167 | Cpeb2 | 296 | Ppp3cb |
| 38 | Pten | 168 | Cpne3 | 297 | Smad5 |
| 39 | Rbbp4 | 169 | Dab2 | 298 | Canx |
| 40 | Rnf13 | 170 | Hpgds | 299 | Ccl2 |
| 41 | Slc25a30 | 171 | Lrrc41 | 300 | Ccl7 |
| 42 | Srsf11 | 172 | Lrrk1 | 301 | Dnm1l |
| 43 | Stard3nl | 173 | Nrk | 302 | Fbn2 |
| 44 | Strn3 | 174 | Pbx1 | 303 | Jmjd1c |
| 45 | Tmem39a | 175 | Prkx | 304 | Nlgn3 |
| 46 | Upk1b | 176 | Rab23 | 305 | Prkacb |
| 47 | AI314180 | 177 | Rbms2 | 306 | Selk |
| 48 | Ccdc66 | 178 | Sec61a2 | 307 | Smurf2 |
| 49 | Chst13 | 179 | Srgap2 | 308 | Srsf1 |
| 50 | Crls1 | 180 | Steap4 | 309 | Ssr1 |
| 51 | Ddit3 | 181 | Taf15 | 310 | Cd209a |
| 52 | Gpc6 | 182 | Tmtc4 | 311 | Dhdh |
| 53 | Hrsp12 | 183 | Usp9× | 312 | Fn1 |
| 54 | Ikzf2 | 184 | Wac | 313 | Hacd3 |
| 55 | Mbnl1 | 185 | Actr2 | 314 | Nbeal1 |
| 56 | Nit2 | 186 | Cdk6 | 315 | Ncaph2 |
| 57 | Npr3 | 187 | Cflar | 316 | Ss18 |
| 58 | Pja2 | 188 | Ddi2 | 317 | Zfp266 |
| 59 | S100pbp | 189 | Dnajc13 | 318 | Zfp36l1 |
| 60 | Siglech | 190 | Dyrk2 | 319 | 4930447C04Rik |
| 61 | Snx27 | 191 | Etnk1 | 320 | Adam10 |
| 62 | Stk3 | 192 | Gadd45a | 321 | Arpc4 |
| 63 | Taok1 | 193 | Khsrp | 322 | Atp8a1 |
| 64 | Tgfbr3 | 194 | Mpdz | 323 | Ddx6 |
| 65 | Tpbg | 195 | N4bp2l2 | 324 | Malat1 |
| 66 | Trak2 | 196 | Otub1 | 325 | Nid1 |
| 67 | Ttc5 | 197 | Rab31 | 326 | Snhg11 |
| 68 | Whsc1l1 | 198 | Sorbs1 | 327 | Cx3cr1 |
| 69 | Zeb2 | 199 | Tacc1 | 328 | Dlk1 |
| 70 | Atrx | 200 | Tmem80 | 329 | Etv5 |
| 71 | Copa | 201 | Tnrc6a | 330 | Mier1 |
| 72 | Cux1 | 202 | Wnt16 | 331 | Rsrc2 |
| 73 | Dmtf1 | 203 | Zcchc7 | 332 | Sdf2l1 |
| 74 | Dnajb6 | 204 | Zdhhc2 | 333 | Slc9a2 |
| 75 | E130308A19Rik | 205 | Zfand5 | 334 | Atic |
| 76 | Epha7 | 206 | Zfp871 | 335 | Camk1d |
| 77 | Fam49a | 207 | Araf | 336 | Gm2a |
| 78 | Gcc2 | 208 | Brd4 | 337 | Mirg |
| 79 | Glg1 | 209 | Cwf19l2 | 338 | Mob1a |
| 80 | Kdelr2 | 210 | Decr2 | 339 | Peli2 |
| 81 | Lnpep | 211 | Hoxb5os | 340 | Pknox1 |
| 82 | Mcf2 | 212 | Hsd17b12 | 341 | Thbs1 |
| 83 | Pcdhb15 | 213 | Krit1 | 342 | Trove2 |
| 84 | Pum1 | 214 | Larp4 | 343 | Dyrk1a |
| 85 | Rhou | 215 | Mapre1 | 344 | Efnb2 |
| 86 | Ssbp2 | 216 | Phip | 345 | Fgfr2 |
| 87 | Stoml2 | 217 | Rbms3 | 346 | Gmfb |
| 88 | Syne2 | 218 | Rhobtb3 | 347 | Lcor |
| 89 | Sypl | 219 | Tspyl1 | 348 | Slc16a6 |
| 90 | Tet1 | 220 | Usf1 | 349 | Sox6 |
| 91 | Tfpi | 221 | Apool | 350 | Srrm2 |
| 92 | Tmed7 | 222 | Ascc3 | 351 | Ugt8a |
| 93 | Trp53 | 223 | Cask | 352 | Arhgef11 |
| 94 | Usp4 | 224 | Cd247 | 353 | Braf |
| 95 | Ywhaz | 225 | Cdh11 | 354 | Dhcr24 |
| 96 | Zbtb20 | 226 | Cdkn2c | 355 | Golt1b |
| 97 | Zcchc3 | 227 | Cpxm1 | 356 | Otud4 |
| 98 | 1810014B01Rik | 228 | Cyp20a1 | 357 | Pfdn2 |
| 99 | Adamts15 | 229 | Ercc4 | 358 | Saa2 |
| 100 | Aplnr | 230 | Fbxl3 | 359 | Smg1 |
| 101 | Arl8a | 231 | Ggact | 360 | Igf2 |
| 102 | Btbd7 | 232 | Hnrnpl | 361 | Lsm12 |
| 103 | Crim1 | 233 | Hook3 | 362 | Malt1 |
| 104 | Fubp1 | 234 | Kantr | 363 | Sc5d |
| 105 | Kansl1 | 235 | Msi2 | 364 | Tial1 |
| 106 | LOC552902 | 236 | Rab14 | 365 | Zfp451 |
| 107 | Lpp | 237 | Rbm12b1 | 366 | Cxadr |
| 108 | Mau2 | 238 | Smek1 | 367 | 9430020K01Rik |
| 109 | Mmgt1 | 239 | Vti1a | 368 | Gpc3 |
| 110 | Mtpn | 240 | 1300017J02Rik | 369 | Serinc3 |
| 111 | Ntrk2 | 241 | E030016H06Rik | 370 | E230029C05Rik |
| 112 | Nudcd1 | 242 | Eif4ebp2 | 371 | Rbm5 |
| 113 | Peak1 | 243 | Gid4 | 372 | Tgfbr1 |
| 114 | Phf14 | 244 | Hsd17b7 | 373 | Yes1 |
| 115 | Polh | 245 | Kat2b | 374 | Vcan |
| 116 | Ppm1k | 246 | Kif1b | 375 | Irf2bp2 |
| 117 | Psmb2 | 247 | Map3k2 | 376 | Dapl1 |
| 118 | Rrm2b | 248 | Nek4 | 377 | Pdia4 |
| 119 | Sfpq | 249 | Ppig | 378 | Uba6 |
| 120 | Sgcd | 250 | Rbm41 | 379 | Wif1 |
| 121 | Sp3 | 251 | Tmem45a | 380 | Slc22a8 |
| 122 | Srpx | 252 | Vamp3 | 381 | Ubfd1 |
| 123 | Stxbp4 | 253 | March7 | 382 | Eya1 |
| 124 | Tcf7l2 | 254 | 9430053O09Rik | 383 | Slc38a5 |
| 125 | Vps13b | 255 | Abi2 | 384 | Nnat |
| 126 | Wnt5a | 256 | Arf6 | 385 | Asgr1 |
| 127 | Zcchc24 | 257 | Arglu1 | 386 | Ranbp3l |
| 128 | Zfp207 | 258 | Cspg5 | 387 | Sall1 |
| 129 | Zkscan8 | 259 | Cybb | 388 | Fabp7 |
| 130 | 5830407P18Rik |
|
|
|
|
 | Table SIII.List of genes specific to type 2
diabetes mellitus. |
Table SIII.
List of genes specific to type 2
diabetes mellitus.
| Gene count | Gene symbol | Gene count | Gene symbol | Gene count | Gene symbol |
|---|
| Upregulated |
|
1 | Sept2 | 242 | Alcam | 48 | Bex1 |
|
2 | Tph2 | 243 | Duoxa1 | 484 | Cables1 |
|
3 | Lipf | 244 | Lrrc3 | 485 | Ccl4 |
|
4 | Ighm | 245 | Tcerg1l | 486 | Cnr2 |
|
5 | Trdn | 246 | Irx3 | 487 | Epgn |
|
6 | Prr32 | 247 | Lep | 488 | Lpcat3 |
|
7 | Oxtr | 248 | Slc16a10 | 489 | Nabp1 |
|
8 | Cpa2 | 249 | Clspn | 490 | Slc24a3 |
|
9 | Gpr50 | 250 | Ctcflos | 491 | Tnfrsf23 |
| 10 | Mbnl3 | 251 | Uhrf1 | 492 | Bcl2l13 |
| 11 | Chil3 | 252 | Ccl2 | 493 | Cd320 |
| 12 | Ucp1 | 253 | Stc2 | 494 | Paqr9 |
| 13 | Ubd | 254 | Gm6484 | 495 | Pih1d2 |
| 14 | Atp1a4 | 255 | Trav9d-3 | 496 | Tmem252 |
| 15 | Ighg | 256 | Gm7694 | 497 | Zwilch |
| 16 | Dusp9 | 257 | Iqgap3 | 498 | 1700013H16Rik |
| 17 | Cdkl4 | 258 | Otop1 | 499 | Dsg1b |
| 18 | Hmga2 | 259 | Ocstamp | 500 | Fabp5 |
| 19 | Atp6v0d2 | 260 | Mylk | 501 | Rhod |
| 20 | Tfr2 | 261 | Ube2t | 502 | Trim25 |
| 21 | Gabrr2 | 262 | C3ar1 | 503 | Wfdc21 |
| 22 | 2010309G21Rik | 263 | Dhx32 | 504 | Hist1h1e |
| 23 | Mmp12 | 264 | Cd200r1 | 505 | Lman1 |
| 24 | Il1rn | 265 | Ccl22 | 506 | Rtn2 |
| 25 | 9030619P08Rik | 266 | Ghrh | 507 | Hoxc13 |
| 26 | Arg1 | 267 | A530020G20Rik | 508 | Krt222 |
| 27 | Olr1 | 268 | A530053G22Rik | 509 | Pfkl |
| 28 | Slc5a7 | 269 | Aifm2 | 510 | Pik3ap1 |
| 29 | Ptchd4 | 270 | 4933427G17Rik | 511 | Sh3tc1 |
| 30 | F7 | 271 | Lat2 | 512 | Srpx2 |
| 31 | Il7r | 272 | Prkag3 | 513 | Tmem120b |
| 32 | Apoc4 | 273 | Ddias | 514 | Tmem144 |
| 33 | Spc25 | 274 | Dgat2 | 515 | Card11 |
| 34 | Mogat2 | 275 | Trim67 | 516 | Etf1 |
| 35 | Lppr4 | 276 | Cenpe | 517 | Plet1os |
| 36 | Crisp2 | 277 | Cd164l2 | 518 | Cd68 |
| 37 | Npr3 | 278 | Mest | 519 | Cdca2 |
| 38 | Rnase2a | 279 | Mgll | 520 | Ccnd2 |
| 39 | Rgs1 | 280 | Slc8a1 | 521 | Dcp2 |
| 40 | 4930502E18Rik | 281 | Gdf3 | 522 | Fgd4 |
| 41 | Klf14 | 282 | Vsig4 | 523 | Foxm1 |
| 42 | Crtac1 | 283 | Adora1 | 524 | Gm6377 |
| 43 | Synpo2 | 284 | Il13ra2 | 525 | Gna13 |
| 44 | Itgax | 285 | Tmem37 | 526 | Il2ra |
| 45 | Sez6l | 286 | Cd1d1 | 527 | Llgl2 |
| 46 | Cdca7l | 287 | Kif2c | 528 | Nckap1l |
| 47 | Serpine1 | 288 | Rrm2 | 529 | Serpina3m |
| 48 | Hmmr | 289 | Cks2 | 530 | Snta1 |
| 49 | Iglv1 | 290 | Pdcd1lg2 | 531 | Hpdl |
| 50 | Fam110c | 291 | Ffar2 | 532 | Knstrn |
| 51 | Kcnj14 | 292 | Stap1 | 533 | Nedd1 |
| 52 | Prr16 | 293 | Pm20d1 | 534 | Nf2 |
| 53 | Cldn23 | 294 | Clec4a2 | 535 | Pitpnm2 |
| 54 | Cyp2r1 | 295 | Fmr1nb | 536 | Pole |
| 55 | Mc2r | 296 | Blnk | 537 | Thbs2 |
| 56 | Grtp1 | 297 | Tnfrsf12a | 538 | Cd300lb |
| 57 | 4930480G23Rik | 298 | Vegfc | 539 | Cdc42ep1 |
| 58 | Dcst1 | 299 | Acaa1b | 540 | Gm30173 |
| 59 | Lpgat1 | 300 | Als2cr12 | 541 | Igfals |
| 60 | Aadac | 301 | Car9 | 542 | Kat7 |
| 61 | 2310002L09Rik | 302 | Gpt | 543 | Kcnn4 |
| 62 | Cyp2c70 | 303 | Havcr2 | 544 | Pmch |
| 63 | Nek2 | 304 | Marco | 545 | Fstl3 |
| 64 | Dnajb13 | 305 | 4931406C07Rik | 546 | Car4 |
| 65 | Gsdma2 | 306 | Dnmt3a | 547 | Cd180 |
| 66 | Zranb3 | 307 | Pik3r5 | 548 | Comt |
| 67 | Orm2 | 308 | Abcc3 | 549 | Fsd2 |
| 68 | Krt79 | 309 | Fbp2 | 550 | Itgb1 |
| 69 | Cxcl2 | 310 | Gm15498 | 551 | Plk4 |
| 70 | Cdca5 | 311 | Kif1b | 552 | Spc24 |
| 71 | Adam8 | 312 | Lpxn | 553 | Tns1 |
| 72 | Tcfl5 | 313 | Pmepa1 | 554 | Atp2a3 |
| 73 | Bub1 | 314 | Slc16a12 | 555 | Myo1f |
| 74 | Agtr2 | 315 | Stil | 556 | Pla2g12a |
| 75 | Dcstamp | 316 | Nup210 | 557 | 1700017B05Rik |
| 76 | Myl4 | 317 | Gdf5 | 558 | Pou2af1 |
| 77 | C430042M11Rik | 318 | Serpina1b | 559 | Rpl39l |
| 78 | Stra6l | 319 | Bche | 560 | A930018P22Rik |
| 79 | Tshr | 320 | Ncaph | 561 | Gm15972 |
| 80 | S100a8 | 321 | Stom | 562 | Usp29 |
| 81 | Ncan | 322 | Hist1h3d | 563 | Arl4a |
| 82 | S100g | 323 | Tmem178 | 564 | Ercc6l |
| 83 | Kcnj15 | 324 | 2610020C07Rik | 565 | Ccl7 |
| 84 | Pkp2 | 325 | Rps6ka3 | 566 | Shmt1 |
| 85 | 1600025M17Rik | 326 | Fam20c | 567 | Atp10a |
| 86 | Gas2l2 | 327 | Trabd2b | 568 | Glb1l2 |
| 87 | D830036C21Rik | 328 | Asf1b | 569 | Slc2a10 |
| 88 | Slc37a2 | 329 | Edem1 | 570 | Smoc1 |
| 89 | Ccnb2 | 330 | Cdt1 | 571 | Zfp106 |
| 90 | Angptl3 | 331 | Cenpa | 572 | Arfgef2 |
| 91 | Htra3 | 332 | AF251705 | 573 | Gcnt1 |
| 92 | Gpnmb | 333 | Hipk1 | 574 | Hesx1 |
| 93 | Cldn15 | 334 | Hn1l | 575 | Mrc2 |
| 94 | Cep55 | 335 | Nepn | 576 | Neurl2 |
| 95 | Abcb4 | 336 | Sel1l | 577 | Plk1 |
| 96 | Gprin3 | 337 | Cyp4b1 | 578 | Zfp426 |
| 97 | Car6 | 338 | Muc13 | 579 | AI844685 |
| 98 | Brca1 | 339 | Rhof | 580 | Ckap2l |
| 99 | Igkv15-103 | 340 | Mcm10 | 581 | Clip1 |
| 100 | Tinag | 341 | Tnfrsf11a | 582 | Dyrk2 |
| 101 | Trim29 | 342 | Ppih | 583 | E2f7 |
| 102 | Cela1 | 343 | Rasgrp2 | 584 | Pex16 |
| 103 | Mki67 | 344 | Srp54b | 585 | Qpctl |
| 104 | Cdk1 | 345 | Gzme | 586 | Slc22a4 |
| 105 | Lrrc39 | 346 | Fam126b | 587 | Tmem132a |
| 106 | Dtl | 347 | Heph | 588 | Adam23 |
| 107 | Ugt3a2 | 348 | Cd200r4 | 589 | D17H6S56E-5 |
| 108 | Dusp3 | 349 | Cnnm2 | 590 | Ece2 |
| 109 | Tmem182 | 350 | Ghr | 591 | Lipa |
| 110 | Acoxl | 351 | Cenpw | 592 | Plek |
| 111 | Chrna2 | 352 | Mcm5 | 593 | Tgfb1 |
| 112 | Apoc2 | 353 | Ska3 | 594 | Adamts9 |
| 113 | Fzd4 | 354 | Mrgprg | 595 | Fgfrl1 |
| 114 | Fads3 | 355 | Pcdh7 | 596 | Mutyh |
| 115 | Arhgap25 | 356 | 2810408I11Rik | 597 | Nim1k |
| 116 | Dock8 | 357 | E2f8 | 598 | Ptprk |
| 117 | Ccnb1 | 358 | 5430437J10Rik | 599 | 5830403F22Rik |
| 118 | Gm6277 | 359 | Kif18b | 600 | Aurka |
| 119 | 4930486L24Rik | 360 | Akr1c13 | 601 | Fignl1 |
| 120 | Hephl1 | 361 | Cd300a | 602 | Isoc1 |
| 121 | Tpx2 | 362 | Cd84 | 603 | Lss |
| 122 | Slpi | 363 | Lrrc27 | 604 | Tgm2 |
| 123 | Sirpb1a | 364 | Mcemp1 | 605 | Trappc13 |
| 124 | Tlr13 | 365 | Adam12 | 606 | Wipi1 |
| 125 | Saa3 | 366 | Ckap2 | 607 | Copg2os2 |
| 126 | 9130204L05Rik | 367 | Fam101b | 608 | Hvcn1 |
| 127 | Cfi | 368 | Igsf6 | 609 | Mpeg1 |
| 128 | Rbm28 | 369 | Pramef12 | 610 | Sphk1 |
| 129 | Dnmt3l | 370 | Tnfrsf22 | 611 | Tpsab1 |
| 130 | Sucnr1 | 371 | Ttk | 612 | Rab20 |
| 131 | Atp6v0a1 | 372 | Car12 | 613 | Skil |
| 132 | Irak2 | 373 | Depdc1a | 614 | Gatb |
| 133 | Prss27 | 374 | Fam64a | 615 | Gm11827 |
| 134 | Birc5 | 375 | Hpgd | 616 | Ptpn22 |
| 135 | Mis18bp1 | 376 | Dbf4 | 617 | Stx17 |
| 136 | Spaca1 | 377 | Igsf21 | 618 | Trib3 |
| 137 | Adh6b | 378 | Kcna5 | 619 | E2f2 |
| 138 | Afp | 379 | AI481207 | 620 | Itsn1 |
| 139 | Tk1 | 380 | Cenph | 621 | C2cd2 |
| 140 | Cxcl3 | 381 | Gla | 622 | Cd44 |
| 141 | Sdsl | 382 | Hrct1 | 623 | Hfe |
| 142 | Tsc22d2 | 383 | Naip1 | 624 | Mmp19 |
| 143 | Klk1 | 384 | Slc18a1 | 625 | 2500002B13Rik |
| 144 | Fosl1 | 385 | 2210406H18Rik | 626 | 2700099C18Rik |
| 145 | Rnf128 | 386 | 3930402G23Rik | 627 | Ano9 |
| 146 | Cdca8 | 387 | Adap2 | 628 | Ccne1 |
| 147 | Timp4 | 388 | Palmd | 629 | Lox |
| 148 | Zc3h12d | 389 | Vldlr | 630 | Dynlrb2 |
| 149 | Plin5 | 390 | Mfge8 | 631 | Eny2 |
| 150 | Ttc39b | 391 | Mpp7 | 632 | Gm3912 |
| 151 | AA467197 | 392 | Sgol1 | 633 | Lipg |
| 152 | Tenm4 | 393 | Mmp13 | 634 | Maff |
| 153 | Nuf2 | 394 | Ms4a6d | 635 | Milr1 |
| 154 | 8430408G22Rik | 395 | Tatdn2 | 636 | Tmem82 |
| 155 | Cdca3 | 396 | Pigt | 637 | 2600006K01Rik |
| 156 | Hilpda | 397 | Sbk1 | 638 | Acvr1c |
| 157 | Esco2 | 398 | Lrg1 | 639 | Bcl2l10 |
| 158 | Slc9a6 | 399 | Oit3 | 640 | Itih5 |
| 159 | Trem2 | 400 | Qsox1 | 641 | Plekhm1 |
| 160 | Pkp1 | 401 | Vwa8 | 642 | Slc7a8 |
| 161 | Tex33 | 402 | Atp8b4 | 643 | Tes |
| 162 | Aqp7 | 403 | B430306N03Rik | 644 | Cd6 |
| 163 | Fmr1 | 404 | Msr1 | 645 | Gsg2 |
| 164 | Fggy | 405 | Nfe2l2 | 646 | Gm2011 |
| 165 | BE949265 | 406 | Nkd1 | 647 | Incenp |
| 166 | Ube2c | 407 | Ptx3 | 648 | Pold3 |
| 167 | Pbk | 408 | Cd22 | 649 | Bst1 |
| 168 | Wif1 | 409 | Galns | 650 | Optc |
| 169 | Nedd9 | 410 | Afmid | 651 | Smad6 |
| 170 | Igfbp3 | 411 | Cd300lg | 652 | Tmem206 |
| 171 | Slc7a10 | 412 | Ermp1 | 653 | Zfp787 |
| 172 | Spdl1 | 413 | Fcgr4 | 654 | Cdc20 |
| 173 | Tmed5 | 414 | Shcbp1 | 655 | Ripk3 |
| 174 | Slc15a3 | 415 | Knop1 | 656 | Sdcbp2 |
| 175 | Clec4d | 416 | Serpina12 | 657 | Myo19 |
| 176 | Serpinb9b | 417 | Larp1b | 658 | Tinagl1 |
| 177 | Cux2 | 418 | Thbs1 | 659 | Tmem106a |
| 178 | Cdk18 | 419 | Seh1l | 660 | Trim16 |
| 179 | C6 | 420 | Lgals3 | 661 | 5033430I15Rik |
| 180 | Lhfpl2 | 421 | Nek6 | 662 | AI606473 |
| 181 | Scel | 422 | Ctse | 663 | Dolk |
| 182 | Glyctk | 423 | Hsd11b2 | 664 | Laptm5 |
| 183 | Mms22l | 424 | Itgb2 | 665 | Lats2 |
| 184 | Tcp11×2 | 425 | Slamf7 | 666 | Lbp |
| 185 | Cacna1e | 426 | 3110027N22Rik | 667 | Ormdl3 |
| 186 | Hipk2 | 427 | Dmrt2 | 668 | Pde1b |
| 187 | Ccl3 | 428 | Hspb6 | 669 | Prelp |
| 188 | Fabp12 | 429 | Ptbp3 | 670 | Serpina3c |
| 189 | Ncapg | 430 | Slc22a12 | 671 | Hr |
| 190 | Pnpla2 | 431 | Ttc7 | 672 | Klhl25 |
| 191 | Dppa3 | 432 | Cenpi | 673 | Shb |
| 192 | Elovl3 | 433 | Cxcl9 | 674 | Vav1 |
| 193 | Prss46 | 434 | Itga7 | 675 | 6030422H21Rik |
| 194 | Mmp8 | 435 | Lcn2 | 676 | Ccdc86 |
| 195 | Folh1 | 436 | Pid1 | 677 | Cenpp |
| 196 | Cenpf | 437 | Prc1 | 678 | Gch1 |
| 197 | Npy | 438 | 1700112E06Rik | 679 | Ms4a6b |
| 198 | Apol6 | 439 | 2010003K11Rik | 680 | Nlrc3 |
| 199 | Rrbp1 | 440 | Plekhg6 | 681 | Gm9899 |
| 200 | Pex5l | 441 | Kif22 | 682 | Phtf2 |
| 201 | F10 | 442 | Ndc80 | 683 | Tead4 |
| 202 | Ccna2 | 443 | Nlrp10 | 684 | Tfcp2l1 |
| 203 | Tmem135 | 444 | Ptges3l | 685 | Tfpi2 |
| 204 | Aspm | 445 | Sod3 | 686 | Map2k5 |
| 205 | Gm8350 | 446 | Abhd15 | 687 | Sh2b2 |
| 206 | Nus1 | 447 | Acsl1 | 688 | Spint2 |
| 207 | Tm4sf5 | 448 | Ltc4s | 689 | Wdr62 |
| 208 | Cdr2 | 449 | Crk | 690 | 4930511E03Rik |
| 209 | Aurkb | 450 | Naip2 | 691 | B230307C23Rik |
| 210 | Lonrf3 | 451 | Peg3 | 692 | Cib2 |
| 211 | Bcar3 | 452 | 4430402I18Rik | 693 | Olfm2 |
| 212 | Dsg1a | 453 | Fgf21 | 694 | Zfp655 |
| 213 | Peg10 | 454 | Lrrc28 | 695 | Cenpk |
| 214 | Tmem38b | 455 | Plekhf2 | 696 | Dgkd |
| 215 | Clec4e | 456 | Rad51ap1 | 697 | Hspa12a |
| 216 | Galnt6 | 457 | Sgol2a | 698 | Hspg2 |
| 217 | Gys2 | 458 | Ehd2 | 699 | Morn5 |
| 218 | Melk | 459 | St8sia4 | 700 | Rbp7 |
| 219 | Spag5 | 460 | Dcbld1 | 701 | Chpt1 |
| 220 | Gsdma | 461 | Mtor | 702 | Ehd3 |
| 221 | Glipr1 | 462 | Plaur | 703 | Hk3 |
| 222 | Pde7a | 463 | Akap2 | 704 | LOC545086 |
| 223 | Top2a | 464 | Ccnf | 705 | Mta3 |
| 224 | Rab38 | 465 | Lrr1 | 706 | Spry4 |
| 225 | Adh4 | 466 | Nat1 | 707 | Treml1 |
| 226 | Cxcl10 | 467 | Gm31718 | 708 | Gpr65 |
| 227 | Kif11 | 468 | Kif20a | 709 | Rel |
| 228 | Hrasls | 469 | Optn | 710 | Stk17b |
| 229 | Kcnk3 | 470 | Plod1 | 711 | Zfp69 |
| 230 | 2810417H13Rik | 471 | Rad54b | 712 | Ccr7 |
| 231 | 1700029I15Rik | 472 | AU021933 | 713 | Cenpn |
| 232 | Serpina3a | 473 | Ccrl2 | 714 | Fbxo42 |
| 233 | Lamb3 | 474 | Piezo1 | 715 | Ttpa |
| 234 | Plet1 | 475 | Stab2 | 716 | Vac14 |
| 235 | Klb | 476 | C5ar1 | 717 | Ncf1 |
| 236 | Btc | 477 | Hgf | 718 | Ybx2 |
| 237 | Ccdc125 | 478 | Kbtbd12 | 719 | Cd300ld |
| 238 | Rbpms | 479 | Map7 | 720 | Gk |
| 239 | Mettl24 | 480 | Rad18 | 721 | Xrcc3 |
| 240 | Tnc | 481 | Sdk2 |
|
|
| 241 | Tpcn2 | 482 | Tlr8 |
|
|
| Downregulated |
|
1 | March2 | 366 | Tspan2 | 731 | St6gal2 |
|
2 | March4 | 367 | Ttyh1 | 732 | Sytl2 |
|
3 | March5 | 368 | Unc13a | 733 | Tnfrsf19 |
|
4 | March6 | 369 | Zfp612 | 734 | Agr2 |
|
5 | 2900055J20Rik | 370 | 1110002L01Rik | 735 | Bcan |
|
6 | Bcl11b | 371 | 9330184L24Rik | 736 | C030034E14Rik |
|
7 | Cdk7 | 372 | Aak1 | 737 | Cntn2 |
|
8 | Esr1 | 373 | Adamtsl1 | 738 | Dcdc2a |
|
9 | Gm13446 | 374 | Aif1l | 739 | E130002L11Rik |
| 10 | Gpr85 | 375 | Ank3 | 740 | Fam131a |
| 11 | Hoxb9 | 376 | Atl1 | 741 | Fam167a |
| 12 | Htr7 | 377 | B930095G15Rik | 742 | Fam169a |
| 13 | Kctd14 | 378 | C030039L03Rik | 743 | Gap43 |
| 14 | Mapk4 | 379 | Cacnb3 | 744 | Gng4 |
| 15 | Ncdn | 380 | Calb2 | 745 | Kcnd1 |
| 16 | Osbpl3 | 381 | Ccdc136 | 746 | Krtdap |
| 17 | Pfn2 | 382 | Cdh8 | 747 | Lrrn1 |
| 18 | Pou3f2 | 383 | Clasp2 | 748 | Lysmd4 |
| 19 | Slc14a1 | 384 | Dlg4 | 749 | Mmd2 |
| 20 | Tardbp | 385 | Epb41l4b | 750 | Nacad |
| 21 | 4833423F13Rik | 386 | Fbxo30 | 751 | Necab1 |
| 22 | Angel1 | 387 | Gpr61 | 752 | Nrxn1 |
| 23 | BC030500 | 388 | Itgb3bp | 753 | Nxpe3 |
| 24 | Cacng4 | 389 | Lcat | 754 | Parm1 |
| 25 | Chd9 | 390 | Lect1 | 755 | Ptprt |
| 26 | Cst6 | 391 | Magi1 | 756 | Pvalb |
| 27 | Dpysl3 | 392 | Meg3 | 757 | Rem2 |
| 28 | Ece1 | 393 | Mmp9 | 758 | Rps6ka6 |
| 29 | Eif5a2 | 394 | Neto2 | 759 | Sphkap |
| 30 | Endod1 | 395 | Otud7a | 760 | Syt11 |
| 31 | Epha5 | 396 | Phactr1 | 761 | Th |
| 32 | Evi5 | 397 | Prkcz | 762 | Thy1 |
| 33 | Gjd2 | 398 | Psd2 | 763 | Tmem74 |
| 34 | Gpr88 | 399 | Rnf130 | 764 | Trim37 |
| 35 | Hs6st2 | 400 | Sema3b | 765 | Tspan8 |
| 36 | Htr1d | 401 | Sema4g | 766 | Wipf3 |
| 37 | Ica1l | 402 | Slc2a3 | 767 | Bves |
| 38 | Kalrn | 403 | Smr2 | 768 | Ccdc92 |
| 39 | Kcnmb2 | 404 | Srr | 769 | Elfn1 |
| 40 | Kcnv1 | 405 | Trpm3 | 770 | Erich3 |
| 41 | Mag | 406 | Zfp385b | 771 | Galnt5 |
| 42 | Nxph1 | 407 | A830010M20Rik | 772 | Gca |
| 43 | Osbpl6 | 408 | AI661384 | 773 | Ggt7 |
| 44 | Pax3 | 409 | AI854703 | 774 | Gria1 |
| 45 | Pcdh9 | 410 | AW046457 | 775 | Hoxb5 |
| 46 | Pcp4l1 | 411 | Adam22 | 776 | Kcnh1 |
| 47 | Pter | 412 | B3gat1 | 777 | Kcnip1 |
| 48 | Rgs7 | 413 | Baiap2l1 | 778 | Kcnmb1 |
| 49 | Scmh1 | 414 | C530044C16Rik | 779 | Lrfn5 |
| 50 | Slc1a6 | 415 | Chrnb2 | 780 | Mab21l2 |
| 51 | Smco3 | 416 | Cntnap5a | 781 | Necab3 |
| 52 | Sostdc1 | 417 | Cplx2 | 782 | Nyap1 |
| 53 | Stxbp1 | 418 | Crh | 783 | Sptbn2 |
| 54 | Stxbp5 | 419 | Csdc2 | 784 | Syn1 |
| 55 | Tmem229b | 420 | Ctxn1 | 785 | Tmem151a |
| 56 | Ube2e3 | 421 | D2Ertd282e | 786 | Tmem25 |
| 57 | Upp2 | 422 | Dclk1 | 787 | Tro |
| 58 | Zfp788 | 423 | Eras | 788 | Trpa1 |
| 59 | 5330417C22Rik | 424 | Erich6 | 789 | Agtr1b |
| 60 | A730089K16Rik | 425 | Gm36529 | 790 | Calca |
| 61 | Abhd10 | 426 | Greb1l | 791 | Camta1 |
| 62 | Adgra1 | 427 | Grm4 | 792 | Crmp1 |
| 63 | C79242 | 428 | Hopx | 793 | Fam131b |
| 64 | Camk4 | 429 | Mycn | 794 | Fam171b |
| 65 | Cep97 | 430 | Nalcn | 795 | Gabra2 |
| 66 | Cpsf2 | 431 | Nebl | 796 | Kcnd3 |
| 67 | Cpsf6 | 432 | Nova1 | 797 | Lmo3 |
| 68 | Csmd3 | 433 | Pcp4 | 798 | Map3k9 |
| 69 | Cyfip2 | 434 | Plekha6 | 799 | Mpped2 |
| 70 | Eml2 | 435 | Rab6b | 800 | Ngfr |
| 71 | Fat3 | 436 | Rbfox3 | 801 | Nmnat2 |
| 72 | Fgf18 | 437 | Rbm11 | 802 | Prkar1b |
| 73 | Golga7b | 438 | Rgs8 | 803 | Rims2 |
| 74 | Gpr173 | 439 | Rtn4 | 804 | Rundc3b |
| 75 | Grin1 | 440 | Sarm1 | 805 | Scn4b |
| 76 | Hs3st1 | 441 | Sema3e | 806 | Slc5a5 |
| 77 | Irf4 | 442 | Shisa4 | 807 | Tesc |
| 78 | Klf12 | 443 | Slc6a17 | 808 | Trpv1 |
| 79 | Lancl1 | 444 | Slco1c1 | 809 | Tst |
| 80 | Lrba | 445 | Synm | 810 | Ube2ql1 |
| 81 | Lrrn4cl | 446 | Tubg2 | 811 | Zfp365 |
| 82 | Lrrtm4 | 447 | Wdr17 | 812 | Zfp804a |
| 83 | Nppb | 448 | 9330182L06Rik | 813 | 2700046A07Rik |
| 84 | Paqr5 | 449 | Aifm3 | 814 | AF529169 |
| 85 | Plch1 | 450 | Bhlha9 | 815 | B3galt5 |
| 86 | Prokr2 | 451 | Cd200r3 | 816 | Bean1 |
| 87 | Prr5l | 452 | Cdh1 | 817 | F2rl2 |
| 88 | R3hdm1 | 453 | Chd5 | 818 | Nrsn1 |
| 89 | Runx1t1 | 454 | Dgkg | 819 | Pcsk1 |
| 90 | Sall4 | 455 | E130309D14Rik | 820 | Pcyt1b |
| 91 | Slc29a4 | 456 | Hapln2 | 821 | Rnf157 |
| 92 | Slc35f2 | 457 | Myt1l | 822 | Serpini1 |
| 93 | Trank1 | 458 | Ntsr2 | 823 | Slain1 |
| 94 | Zdhhc18 | 459 | Oprl1 | 824 | Slc35f4 |
| 95 | Zfp318 | 460 | Prdm8 | 825 | Slc4a10 |
| 96 | Zfp37 | 461 | Prr18 | 826 | Vps13c |
| 97 | 2900009J20Rik | 462 | Psmf1 | 827 | Wt1 |
| 98 | A930041H05Rik | 463 | Rab3a | 828 | Adamts16 |
| 99 | Ak5 | 464 | Rasgef1a | 829 | Ano3 |
| 100 | Arg2 | 465 | Rgmb | 830 | Arfgef3 |
| 101 | Bank1 | 466 | Rgs7bp | 831 | Arhgdig |
| 102 | Ccdc28a | 467 | Robo1 | 832 | Cntn1 |
| 103 | Cdkn1b | 468 | Rph3a | 833 | Ddx25 |
| 104 | Clec2l | 469 | Slc2a12 | 834 | Dzank1 |
| 105 | Csmd1 | 470 | Slc6a2 | 835 | Fam19a3 |
| 106 | Dip2a | 471 | Sstr4 | 836 | Gprin2 |
| 107 | Dlgap1 | 472 | Syt7 | 837 | Hsbp1l1 |
| 108 | F5 | 473 | Tdo2 | 838 | Htr4 |
| 109 | Fzd3 | 474 | Tmem108 | 839 | Jakmip1 |
| 110 | Htr1a | 475 | Vwa5a | 840 | Jakmip2 |
| 111 | Hunk | 476 | Zfr2 | 841 | Klc1 |
| 112 | Klhl32 | 477 | 9530077C05Rik | 842 | Lix1 |
| 113 | L1cam | 478 | Acsl6 | 843 | Maneal |
| 114 | Lrrtm2 | 479 | Aqp4 | 844 | Npy2r |
| 115 | Map6d1 | 480 | Atcay | 845 | Rab3c |
| 116 | Mfsd6 | 481 | Ccdc68 | 846 | Reep2 |
| 117 | Mrpl35 | 482 | Cdk5r1 | 847 | Rufy2 |
| 118 | Nedd4l | 483 | Cgref1 | 848 | Sh3gl2 |
| 119 | Pcsk6 | 484 | Cpb1 | 849 | Vstm5 |
| 120 | Plxnb1 | 485 | Ctnnd2 | 850 | 1810041L15Rik |
| 121 | Ppp1r2 | 486 | Cxxc4 | 851 | A830039N20Rik |
| 122 | Rbm33 | 487 | Dnah7a | 852 | Aldoc |
| 123 | Rfx4 | 488 | Dzip3 | 853 | Asic1 |
| 124 | Sacs | 489 | Fndc4 | 854 | BC059841 |
| 125 | Serpina11 | 490 | Fxyd7 | 855 | Car10 |
| 126 | Shd | 491 | Gabra5 | 856 | Ccdc172 |
| 127 | Tcte2 | 492 | Glrb | 857 | Cldn10 |
| 128 | Tia1 | 493 | Lrch1 | 858 | D130043K22Rik |
| 129 | Tox2 | 494 | Mir124a-1hg | 859 | Gabrg2 |
| 130 | Ttbk1 | 495 | Ncam2 | 860 | Gm16551 |
| 131 | Ttc3 | 496 | Plk5 | 861 | Kcnip2 |
| 132 | Vamp4 | 497 | Pogz | 862 | Kcnip4 |
| 133 | 2900056L01Rik | 498 | Prox1 | 863 | Kif3c |
| 134 | 2900064F13Rik | 499 | Rasl10b | 864 | Mrgpra3 |
| 135 | Arhgap20 | 500 | Rnf208 | 865 | Pappa2 |
| 136 | Ccdc112 | 501 | Scin | 866 | Rab39b |
| 137 | Cfap69 | 502 | Scn8a | 867 | Rassf6 |
| 138 | Cmtm5 | 503 | Scrt1 | 868 | Rbfox1 |
| 139 | Dtna | 504 | Slitrk4 | 869 | Rufy3 |
| 140 | Dusp15 | 505 | Susd5 | 870 | Sez6l2 |
| 141 | Ephx4 | 506 | Tmem169 | 871 | Soga3 |
| 142 | Faim2 | 507 | Tmem196 | 872 | Stac |
| 143 | Gabrb3 | 508 | Ttc39c | 873 | Sv2a |
| 144 | Galnt14 | 509 | Tyrp1 | 874 | Tmem179 |
| 145 | Gm12371 | 510 | Unc5c | 875 | Tspyl4 |
| 146 | Gm13889 | 511 | Vwc2l | 876 | Aplp1 |
| 147 | Gprasp1 | 512 | 5930412G12Rik | 877 | Atp6v1g2 |
| 148 | Hspa4l | 513 | Abcg4 | 878 | Far2 |
| 149 | Igsf11 | 514 | Ache | 879 | Megf11 |
| 150 | Itih3 | 515 | Acsm3 | 880 | Nsg2 |
| 151 | Kcnip3 | 516 | Arhgap12 | 881 | Pclo |
| 152 | Kcnk1 | 517 | Cabp7 | 882 | Pou4f2 |
| 153 | Lingo1 | 518 | Ccdc30 | 883 | Ptprn |
| 154 | Mkx | 519 | Clip3 | 884 | Rasgrp1 |
| 155 | Nipa1 | 520 | Clip4 | 885 | Rit2 |
| 156 | Phex | 521 | Dgkb | 886 | Slc1a2 |
| 157 | Pnkd | 522 | Diras1 | 887 | Them5 |
| 158 | Ppm1l | 523 | Elovl2 | 888 | Tmem151b |
| 159 | Spink10 | 524 | Gm10700 | 889 | Zwint |
| 160 | St3gal6 | 525 | Gpr149 | 890 | 6430604M11Rik |
| 161 | St6galnac3 | 526 | Has2 | 891 | Bsn |
| 162 | Stk33 | 527 | Hoxd10 | 892 | C530008M17Rik |
| 163 | Tfap2a | 528 | Hs3st4 | 893 | Cadps2 |
| 164 | Wdr31 | 529 | Iqsec3 | 894 | Cbln2 |
| 165 | 4930519N06Rik | 530 | Kazn | 895 | Ctnna2 |
| 166 | Acsbg1 | 531 | Kcnf1 | 896 | Cyp4×1 |
| 167 | Akr1c18 | 532 | Kif3a | 897 | Elavl3 |
| 168 | Asxl3 | 533 | Lepr | 898 | Fbxo2 |
| 169 | Atad1 | 534 | Lingo2 | 899 | Gm10419 |
| 170 | B3galt1 | 535 | Mapt | 900 | Igfbp2 |
| 171 | B630019K06Rik | 536 | Mycl | 901 | Jph3 |
| 172 | Ccdc138 | 537 | Neto1 | 902 | Jph4 |
| 173 | Cdr2l | 538 | Nipal3 | 903 | Klhdc8a |
| 174 | Clec3b | 539 | Nwd2 | 904 | Nrcam |
| 175 | Cyr61 | 540 | Pdzd4 | 905 | Ppap2c |
| 176 | Dhh | 541 | Plcxd3 | 906 | Ppm1j |
| 177 | Edil3 | 542 | Plp1 | 907 | Ptgs2 |
| 178 | Fign | 543 | Ppfia2 | 908 | Tcte1 |
| 179 | Gabbr1 | 544 | Ptbp2 | 909 | Tmem150c |
| 180 | Gabra1 | 545 | Scrn1 | 910 | Trim9 |
| 181 | Gm13629 | 546 | Slc25a18 | 911 | Acpp |
| 182 | Gm5089 | 547 | Slco5a1 | 912 | Adgrb3 |
| 183 | Gnaz | 548 | Svop | 913 | Anks1b |
| 184 | Gpr62 | 549 | Syngr1 | 914 | Ccdc184 |
| 185 | Hcn1 | 550 | Tmem246 | 915 | Chga |
| 186 | Kcnk5 | 551 | Tmem88b | 916 | Ckmt1 |
| 187 | Kcnq5 | 552 | Arpp21 | 917 | Galnt13 |
| 188 | Lrrc49 | 553 | Atp1b1 | 918 | Gng3 |
| 189 | Mbp | 554 | Cd209g | 919 | Kcnab1 |
| 190 | Pak1 | 555 | Cda | 920 | Kcnc1 |
| 191 | Pax9 | 556 | Chst5 | 921 | Kif5a |
| 192 | Pcdh10 | 557 | Cnnm1 | 922 | Napb |
| 193 | Praf2 | 558 | Dleu2 | 923 | Nrxn3 |
| 194 | Prss12 | 559 | Dscam | 924 | Ptgds |
| 195 | Ptchd2 | 560 | Dusp26 | 925 | Ptprn2 |
| 196 | Rimklb | 561 | Hebp2 | 926 | Rgs17 |
| 197 | Sgtb | 562 | Il31ra | 927 | Tox3 |
| 198 | Sh3d19 | 563 | Lhfpl3 | 928 | Uchl1 |
| 199 | Slc16a9 | 564 | Lrp8 | 929 | Zmat4 |
| 200 | Smpd3 | 565 | Mfsd4 | 930 | Csrnp3 |
| 201 | Sorcs3 | 566 | P2rx3 | 931 | Dpp6 |
| 202 | Stmn2 | 567 | Pigz | 932 | Fam184b |
| 203 | Usp31 | 568 | Pnma2 | 933 | Gdap1 |
| 204 | Vamp1 | 569 | Rab39 | 934 | Kctd8 |
| 205 | Vat1l | 570 | Rell2 | 935 | Nrg3 |
| 206 | Zdbf2 | 571 | Rlbp1 | 936 | Rnf112 |
| 207 | Aldh1a2 | 572 | Sema4f | 937 | Syt2 |
| 208 | Atp2b4 | 573 | Slitrk5 | 938 | Tmem200c |
| 209 | Cacnb4 | 574 | Smarcal1 | 939 | 3632451O06Rik |
| 210 | Camk1g | 575 | Sntg1 | 940 | 9430021M05Rik |
| 211 | Chn1 | 576 | Stmn3 | 941 | AW060742 |
| 212 | Dio3os | 577 | Susd4 | 942 | Ank1 |
| 213 | Fkbp1b | 578 | Tmem117 | 943 | Asic3 |
| 214 | Gjb1 | 579 | Vwa7 | 944 | Brsk2 |
| 215 | Gna14 | 580 | 1700019D03Rik | 945 | Cldn11 |
| 216 | Herc3 | 581 | A2m | 946 | Faxc |
| 217 | Hnrnpr | 582 | Brsk1 | 947 | Fstl5 |
| 218 | Igsf1 | 583 | C030011L09Rik | 948 | Lhfpl5 |
| 219 | Irf6 | 584 | Cabp1 | 949 | Nptx2 |
| 220 | Ism1 | 585 | Dok4 | 950 | Pacsin1 |
| 221 | Jam3 | 586 | Epb41l3 | 951 | Rims3 |
| 222 | Klf7 | 587 | Exph5 | 952 | Rspo2 |
| 223 | Ltbp1 | 588 | Fez1 | 953 | Rundc3a |
| 224 | Mapk8ip1 | 589 | Fmn2 | 954 | Scg5 |
| 225 | Nek1 | 590 | Gria2 | 955 | Slc6a15 |
| 226 | Nrip3 | 591 | Kif5c | 956 | Srrm3 |
| 227 | Phyhip | 592 | Mast1 | 957 | St8sia3 |
| 228 | Prepl | 593 | Ms4a3 | 958 | Tram1l1 |
| 229 | Rasgrf1 | 594 | Ndrg4 | 959 | Zswim5 |
| 230 | Serpind1 | 595 | Pirt | 960 | 2900011O08Rik |
| 231 | Sox1 | 596 | Prmt8 | 961 | Astn1 |
| 232 | Sox2ot | 597 | Rab33a | 962 | Atp2b2 |
| 233 | Spock2 | 598 | Rab37 | 963 | Bend6 |
| 234 | Susd2 | 599 | Slc25a12 | 964 | Bex2 |
| 235 | Syndig1l | 600 | Srgap3 | 965 | Cacna2d3 |
| 236 | Zfhx2 | 601 | Tmem163 | 966 | Cckar |
| 237 |
A730054J21Rik | 602 | Tmem79 | 967 | Cend1 |
| 238 | Adam11 | 603 | Ttc9b | 968 |
D930028M14Rik |
| 239 | Adcy1 | 604 |
4930524O07Rik | 969 | Dnm3 |
| 240 | Adgrl3 | 605 |
A330050F15Rik | 970 | Fam155a |
| 241 | Ankrd13d | 606 |
A330102I10Rik | 971 | Gnal |
| 242 | Apba1 | 607 | Arhgap28 | 972 | Ipw |
| 243 | Apcdd1 | 608 | Arhgap44 | 973 | Kcna4 |
| 244 | B3gnt5 | 609 | Celf4 | 974 | Kcnj10 |
| 245 | Bmp7 | 610 | Cntnap4 | 975 | Lsamp |
| 246 | Crym | 611 | Elovl7 | 976 | Mapk8ip2 |
| 247 | Dbndd2 | 612 | Fcrls | 977 | Mtcl1 |
| 248 | Eef1a2 | 613 | Gpr37 | 978 | Phf24 |
| 249 | Fam81a | 614 | Grin3a | 979 | Snap25 |
| 250 | Fbxl16 | 615 | Ibsp | 980 | Tmem59l |
| 251 | Fbxo41 | 616 | Ints6 | 981 | Atp2b3 |
| 252 | Flrt1 | 617 | Lgi3 | 982 | Col25a1 |
| 253 | Gkn3 | 618 | Lrrtm1 | 983 | Dpysl5 |
| 254 | Hey2 | 619 | Magee1 | 984 | Grip1 |
| 255 | Kcnd2 | 620 | Pcdh20 | 985 | Hapln1 |
| 256 | Kcng4 | 621 | Plcxd2 | 986 | Klk6 |
| 257 | Lin7a | 622 | Plekhh1 | 987 | Lhfpl4 |
| 258 | Lrrtm3 | 623 | Prkg2 | 988 | Nap1l3 |
| 259 | Ncam1 | 624 | Prune2 | 989 | Ppm1e |
| 260 | Nrsn2 | 625 | Rcan2 | 990 | Prph |
| 261 | Pak7 | 626 | Scn2b | 991 | Rnf182 |
| 262 | Pcolce2 | 627 | Slc7a14 | 992 | Rtn1 |
| 263 | Ppp2r2c | 628 | Stx1b | 993 | Slc39a12 |
| 264 | Prr15 | 629 | Sult4a1 | 994 | Tmem35 |
| 265 | S100b | 630 | Syt14 | 995 | Zcchc18 |
| 266 | Samd14 | 631 | Tmem132e | 996 | Ano4 |
| 267 | Sh3gl3 | 632 | Ubash3b | 997 | Arhgef7 |
| 268 | Slc6a11 | 633 |
A830018L16Rik | 998 | BC048546 |
| 269 | Trio | 634 | Adcy8 | 999 | Gabrg1 |
| 270 |
2610100L16Rik | 635 | Cdh10 | 1000 | Hecw1 |
| 271 | AI846148 | 636 | Ceacam10 | 1001 | Insm2 |
| 272 | Apbb1 | 637 | Chrna6 | 1002 | Map7d2 |
| 273 | Ccl27a | 638 | Eno2 | 1003 | Mog |
| 274 | Ccp110 | 639 | Fgf4 | 1004 | Pcsk2 |
| 275 | Cd24a | 640 | Frrs1l | 1005 | Pgbd5 |
| 276 | Clvs2 | 641 | Gm5124 | 1006 | Rprm |
| 277 | Cntn6 | 642 | Gpr22 | 1007 | Sgpp2 |
| 278 | Cxcl13 | 643 | Il1r2 | 1008 | Slc17a7 |
| 279 | Cytl1 | 644 | Lancl3 | 1009 | Slc26a7 |
| 280 | Depdc5 | 645 | Ogfrl1 | 1010 | Spock1 |
| 281 | Dkk2 | 646 | Olfm3 | 1011 | Adrb3 |
| 282 | Gabrb2 | 647 | Phactr3 | 1012 | Akap6 |
| 283 | Gm15663 | 648 | Reep1 | 1013 |
C030017B01Rik |
| 284 | Hs3st2 | 649 | Rgs4 | 1014 | Calb1 |
| 285 | Id4 | 650 | Rimbp2 | 1015 | Cpne4 |
| 286 | Il18 | 651 | Skida1 | 1016 | Dync1i1 |
| 287 | Map1a | 652 | Slc6a1 | 1017 | Isl1 |
| 288 | Mllt11 | 653 | Slitrk1 | 1018 | Isl2 |
| 289 | Nfasc | 654 | Tmem56 | 1019 | Kndc1 |
| 290 | Pak3 | 655 | Trnp1 | 1020 | Map4 |
| 291 | Pgm2l1 | 656 | Tspyl5 | 1021 | Mapk10 |
| 292 | Plcd4 | 657 | Vstm2a | 1022 | Phyhipl |
| 293 | Rasal2 | 658 |
4832406H04Rik | 1023 | Pou4f1 |
| 294 | Rltpr | 659 |
A330068G13Rik | 1024 | Rims1 |
| 295 | Sec62 | 660 | Aard | 1025 | Sv2b |
| 296 | Snph | 661 | Acsl3 | 1026 | Syt1 |
| 297 | Stk32c | 662 | Atp8a2 | 1027 | Syt4 |
| 298 | Syngr3 | 663 | Cacna1b | 1028 | Tubb4a |
| 299 | Tbc1d30 | 664 | Cdh9 | 1029 | Amph |
| 300 | Trim2 | 665 | Ces1f | 1030 | Elavl4 |
| 301 |
1110032F04Rik | 666 | Col4a6 | 1031 | Ermn |
| 302 | Ank2 | 667 | Dner | 1032 | Gfap |
| 303 | Apc2 | 668 | Fn3k | 1033 | Hoxd1 |
| 304 | Apoh | 669 | Gm16532 | 1034 | Mlc1 |
| 305 | Asphd1 | 670 | Itga8 | 1035 | Mup10 |
| 306 | BB319198 | 671 | Kif1a | 1036 | Nrg1 |
| 307 | Cacna2d2 | 672 | L1td1 | 1037 | Scn10a |
| 308 | Cadm3 | 673 | Megf10 | 1038 | Tagln3 |
| 309 | Clgn | 674 | Mtus2 | 1039 | Vgf |
| 310 | Cmah | 675 | Nipal2 | 1040 | Add2 |
| 311 | Ddn | 676 | Nrxn2 | 1041 | Cdhr1 |
| 312 | Dhrs2 | 677 | Ntrk1 | 1042 | Chrna7 |
| 313 | Efcab1 | 678 | Pcdh8 | 1043 | Cpne6 |
| 314 | Fam189a1 | 679 | Pla2g3 | 1044 | Dlg2 |
| 315 | Gm32444 | 680 | Rab9b | 1045 | Dpp10 |
| 316 | Gpr158 | 681 | Sowahb | 1046 | Fam19a4 |
| 317 | Gpr75 | 682 | Trpc3 | 1047 | Grik1 |
| 318 | Grm8 | 683 | Trpc6 | 1048 | Htr3a |
| 319 | Inadl | 684 | Tvp23a | 1049 | Plekhd1 |
| 320 | Kcnq2 | 685 | Vwa5b2 | 1050 | Ppp1r1c |
| 321 | Kctd4 | 686 |
A730017C20Rik | 1051 | Ppp2r2b |
| 322 | Kif21a | 687 | Amer3 | 1052 | Ralyl |
| 323 | Krt27 | 688 | Arhgef4 | 1053 | St8sia1 |
| 324 | Lgi2 | 689 | Arnt2 | 1054 | Stmn4 |
| 325 | Lonrf2 | 690 | Ccser1 | 1055 | Synpr |
| 326 | Lztfl1 | 691 | Cntnap2 | 1056 |
2900052N01Rik |
| 327 | Mfsd2a | 692 | Cplx1 | 1057 | Chgb |
| 328 | Mro | 693 | Crygs | 1058 | Fam19a2 |
| 329 | Pcnxl2 | 694 | Dock3 | 1059 | Resp18 |
| 330 | Prkg1 | 695 | Dpp4 | 1060 | Slc17a6 |
| 331 | Srd5a1 | 696 | Fam189b | 1061 | Spock3 |
| 332 | Srsf12 | 697 | Gdap1l1 | 1062 | Vsnl1 |
| 333 | Tmem231 | 698 | Gm2115 | 1063 | Nefl |
| 334 | Tnik | 699 | Gprasp2 | 1064 | Scn9a |
| 335 | Trim36 | 700 | Hoxb6 | 1065 | Nts |
| 336 | Ttbk2 | 701 | Lgi1 | 1066 | Sult1e1 |
| 337 | Aox3 | 702 | Mobp | 1067 | Avil |
| 338 | Ccdc47 | 703 | Mogat1 | 1068 |
2810037O22Rik |
| 339 | Celsr2 | 704 | Mtmr7 | 1069 | Nefh |
| 340 | Cldn9 | 705 | Nkx2-2 | 1070 | Cdk5r2 |
| 341 | Dbndd1 | 706 | Omg | 1071 | Ttc9 |
| 342 | Esd | 707 | Pde1c | 1072 | Adcyap1 |
| 343 | Fxyd3 | 708 | Ptprz1 | 1073 | Pon1 |
| 344 | Greb1 | 709 | Scn1a | 1074 | Slc24a2 |
| 345 | Gria4 | 710 | Slc35f3 | 1075 | Nefm |
| 346 | Kcnn2 | 711 | Spire2 | 1076 | Panx2 |
| 347 | Kcnt1 | 712 | Stxbp5l | 1077 | Syt9 |
| 348 | Lynx1 | 713 | Tenm1 | 1078 | Rimkla |
| 349 | Ndp | 714 | Timd4 | 1079 | Gm2102 |
| 350 | Nkain2 | 715 |
6330563C09Rik | 1080 | Scg2 |
| 351 | Pi16 | 716 | Ap3b2 | 1081 | Tac1 |
| 352 | Pip5kl1 | 717 | Arhgap36 | 1082 | Prrxl1 |
| 353 | Pla2g2d | 718 | Caln1 | 1083 | AI593442 |
| 354 | Ppl | 719 | Celf6 | 1084 | Calcb |
| 355 | Prokr1 | 720 | Cntn4 | 1085 | Shh |
| 356 | Rab27b | 721 | Corin | 1086 | Tmem130 |
| 357 | Rragd | 722 | Epb41 | 1087 | Cyp2f2 |
| 358 | Rsrp1 | 723 | Fosb | 1088 | Snap91 |
| 359 | Scrt2 | 724 | Kcnj3 | 1089 | Nap1l2 |
| 360 | Slc35d3 | 725 | Kcnk10 | 1090 | Scn11a |
| 361 | Spink2 | 726 | Lrrc75b | 1091 | Myt1 |
| 362 | Stk39 | 727 | Morn4 | 1092 | Grm7 |
| 363 | Synrg | 728 | Onecut2 | 1093 | Tubb3 |
| 364 | Tmem255a | 729 | Pcsk1n | 1094 | Cadps |
| 365 | Tshz2 | 730 | Scg3 |
|
|
GO and pathway enrichment
analyses
There were 60 coincident enriched GO terms for DEGs
in T1- and T2DM, the top 20 of which are listed in Table I. In addition, the top 20 enriched GO
terms for DEGs that were unique to sciatic nerves from the T1- or
T2DM model are presented in Table
II. Besides these, 21 and 27 KEGG pathways were enriched for
DEGs in T1- and T2DM, respectively. Among them, 4 pathways were
coincident in both models (Table
III), while 17 and 23 pathways were unique to T1- and T2DM
respectively (Table IV).
 | Table I.Top 20 coincident enriched GO terms
in T1- and T2DM. |
Table I.
Top 20 coincident enriched GO terms
in T1- and T2DM.
|
|
| T1DM | T2DM |
|---|
|
|
|
|
|
|---|
| GO ID | GO term | Gene count | P-value | Gene count | P-value |
|---|
| GO:0005515
MF | Protein
binding | 57 | 3.1E-6 | 76 | 9.2E-20 |
| GO:0005737
CC | Cytoplasm | 21 | 6.7E-6 | 123 | 3.8E-14 |
| GO:0001525
BP | Angiogenesis | 14 | 9.6E-6 | 89 | 7.3E-18 |
| GO:0000166
MF | Nucleotide
binding | 45 | 2.4E-5 | 46 | 5.5E-10 |
| GO:0007399
BP | Nervous system
development | 21 | 3.2E-5 | 86 | 7.3E-11 |
| GO:0008201
MF | Heparin
binding | 13 | 4.6E-5 | 57 | 5.9E-14 |
| GO:0006468
BP | Protein
phosphorylation | 11 | 5.4E-5 | 61 | 4.3E-12 |
| GO:0005578
CC | Proteinaceous
extracellular matrix | 24 | 5.5E-5 | 66 | 9.1E-20 |
| GO:0016310
BP |
Phosphorylation | 13 | 6.2E-5 | 78 | 8.8E-16 |
| GO:0009986
CC | Cell surface | 12 | 6.5E-5 | 54 | 5.6E-17 |
| GO:0016020
CC | Membrane | 17 | 8.9E-5 | 23 | 3.2E-15 |
| GO:0005794
CC | Golgi
apparatus | 12 | 2.4E-4 | 45 | 2.5E-17 |
| GO:0004672
MF | Protein kinase
activity | 14 | 3.1E-4 | 24 | 4.2E-16 |
| GO:0030335
BP | Positive
regulation of cell migration | 15 | 4.5E-4 | 19 | 8.5E-17 |
| GO:0005783
CC | Endoplasmic
reticulum |
6 | 5.3E-4 | 32 | 5.2E-16 |
| GO:0019933
BP | cAMP-mediated
signaling | 18 | 5.8E-4 | 30 | 7.5E-13 |
| GO:0048471
CC | Perinuclear
region of cytoplasm | 16 | 6.3E-4 | 21 | 5.2E-12 |
| GO:0016301
MF | Kinase
activity | 19 | 6.7E-4 | 26 | 7.1E-15 |
| GO:0004674
MF | Protein
serine/threonine kinase activity | 32 | 1.3E-3 | 24 | 2.1E-18 |
| GO:0006629
BP | Lipid metabolic
process | 22 | 1.6E-3 | 42 | 7.6E-22 |
 | Table II.Top 20 enriched GO terms for
differentially expressed genes in the sciatic nerve of T1- and T2DM
models, respectively. |
Table II.
Top 20 enriched GO terms for
differentially expressed genes in the sciatic nerve of T1- and T2DM
models, respectively.
| GO ID | GO term | Gene count | P-value |
|---|
| T1DM |
| GO:0005654
CC | Nucleoplasm | 92 | 5.3E-7 |
| GO:0070062
CC | Extracellular
exosome | 116 | 1.3E-6 |
| GO:0005829
CC | Cytosol | 84 | 2.8E-6 |
| GO:0045893
BP | Positive
regulation of transcription, DNA-templated | 38 | 4.3E-6 |
| GO:0050680
BP | Negative
regulation of epithelial cell proliferation | 12 | 6.0E-6 |
| GO:0071560
BP | Cellular response
to transforming growth factor β stimulus | 11 | 8.3E-6 |
| GO:0005604
CC | Basement
membrane | 13 | 1.7E-5 |
| GO:0090090
BP | Negative
regulation of canonical Wnt signaling pathway | 13 | 3.5E-5 |
| GO:0045165
BP | Cell fate
commitment | 11 | 3.7E-5 |
| GO:0005925
CC | Focal
adhesion | 27 | 4.7E-5 |
| GO:0030154
BP | Cell
differentiation | 43 | 6.9E-5 |
| GO:0045944
BP | Positive
regulation of transcription from RNA polymerase II promoter | 50 | 1.6E-4 |
| GO:0005634
CC | Nucleus | 208 | 2.4E-4 |
| GO:0009887
BP | Organ
morphogenesis | 12 | 2.9E-4 |
| GO:0035925
MF | mRNA 3′-UTR
AU-rich region binding | 5 | 3.0E-4 |
| GO:0008285
BP | Negative
regulation of cell proliferation | 25 | 3.1E-4 |
| GO:0048709
BP | Oligodendrocyte
differentiation | 7 | 3.6E-4 |
| GO:0000381
BP | Regulation of
alternative mRNA splicing, via spliceosome | 7 | 7.8E-4 |
| GO:0000122
BP | Negative
regulation of transcription from RNA polymerase II promoter | 37 | 1.1E-3 |
| GO:0031012
CC | Extracellular
matrix | 19 | 1.7E-3 |
| T2DM |
| GO:0030054
CC | Cell
junction | 147 | 1.4E-26 |
| GO:0030424
CC | Axon | 98 | 1.9E-26 |
| GO:0043195
CC | Terminal
bouton | 46 | 8.7E-21 |
| GO:0006811
BP | Ion
transport | 112 | 5.1E-18 |
| GO:0005216
MF | Ion channel
activity | 48 | 1.8E-14 |
| GO:0042734
CC | Presynaptic
membrane | 31 | 2.4E-14 |
| GO:0034765
BP | Regulation of ion
transmembrane transport | 42 | 3.8E-14 |
| GO:0008021
CC | Synaptic
vesicle | 39 | 3.6E-13 |
| GO:0005244
MF | Voltage-gated ion
channel activity | 40 | 5.0E-13 |
| GO:0005509
MF | Calcium ion
binding | 111 | 2.9E-12 |
| GO:0045211
CC | Postsynaptic
membrane | 54 | 3.6E-13 |
| GO:0005886
CC | Plasma
membrane | 509 | 3.5E-12 |
| GO:0006810
BP | Transport | 227 | 1.1E-11 |
| GO:0007268
BP | Chemical synaptic
transmission | 44 | 1.4E-11 |
| GO:0008076
CC | Voltage-gated
potassium channel complex | 27 | 2.2E-10 |
| GO:0007269
BP | Neurotransmitter
secretion | 17 | 3.6E-11 |
| GO:0000775
CC | Chromosome,
centromeric region | 38 | 7.1E-11 |
| GO:0006813
BP | Potassium ion
transport | 34 | 1.1E-9 |
| GO:0005267
MF | Potassium channel
activity | 26 | 1.4E-9 |
| GO:0005887
CC | Integral
component of plasma membrane | 150 | 5.7E-10 |
 | Table III.Coincident enriched Kyoto
Encyclopedia of Genes and Genomes pathways in T1- and T2DM. |
Table III.
Coincident enriched Kyoto
Encyclopedia of Genes and Genomes pathways in T1- and T2DM.
|
| T1DM | T2DM |
|---|
|
|
|
|
|---|
| Term | Gene count | P-value | Gene count | P-value |
|---|
| mmu04010:MAPK
signaling pathway | 19 | 7.5E-4 | 29 | 3.2E-2 |
|
mmu04512:ECM-receptor interaction | 10 | 1.4E-3 | 15 | 6.9E-3 |
| mmu04390:Hippo
signaling pathway | 11 | 1.8E-2 | 24 | 1.2E-3 |
| mmu04360:Axon
guidance |
9 | 4.5E-2 | 17 | 3.8E-2 |
 | Table IV.Enriched Kyoto Encyclopedia of Genes
and Genomes pathways in T1- and T2DM, respectively. |
Table IV.
Enriched Kyoto Encyclopedia of Genes
and Genomes pathways in T1- and T2DM, respectively.
| Term | Gene count | P-value |
|---|
| T1DM |
| mmu04010:MAPK
signaling pathway | 19 | 7.5E-4 |
|
mmu04512:ECM-receptor interaction | 10 | 1.4E-3 |
|
mmu05202:Transcriptionalmisregulation in
cancer | 14 | 1.7E-3 |
| mmu05200:Pathways
in cancer | 24 | 2.5E-3 |
|
mmu05205:Proteoglycans in cancer | 15 | 3.8E-3 |
| mmu04310:Wnt
signaling pathway | 11 | 1.1E-2 |
|
mmu05214:Glioma | 7 | 1.4E-2 |
| mmu05222:Small
cell lung cancer | 8 | 1.4E-2 |
| mmu04015:Rap1
signaling pathway | 14 | 1.4E-2 |
| mmu04330:Notch
signaling pathway | 6 | 1.7E-2 |
|
mmu04914:Progesterone-mediated | 8 | 1.7E-2 |
| oocyte
maturation |
| mmu04390:Hippo
signaling pathway | 11 | 1.8E-2 |
| mmu00100:Steroid
biosynthesis | 4 | 1.9E-2 |
|
mmu05218:Melanoma | 7 | 2.1E-2 |
|
mmu04614:Renin-angiotensin system | 5 | 2.2E-2 |
| mmu04520:Adherens
junction | 7 | 2.3E-2 |
| mmu04510:Focal
adhesion | 13 | 2.5E-2 |
| mmu04913:Ovarian
steroidogenesis | 6 | 3.0E-2 |
| mmu04360:Axon
guidance | 9 | 4.5E-2 |
|
mmu05210:Colorectal cancer | 6 | 4.6E-2 |
|
mmu05212:Pancreatic cancer | 6 | 4.9E-2 |
| T2DM |
| mmu05033:Nicotine
addiction | 19 | 1.6E-10 |
|
mmu04080:Neuroactive ligand-receptor
interaction | 49 | 1.4E-7 |
|
mmu04723:Retrograde endocannabinoid
signaling | 23 | 8.5E-6 |
| mmu04721:Synaptic
vesicle cycle | 17 | 1.2E-5 |
| mmu05032:Morphine
addiction | 20 | 6.7E-5 |
| mmu04514:Cell
adhesion molecules (CAMs) | 27 | 2.4E-4 |
|
mmu04724:Glutamatergic synapse | 21 | 4.3E-4 |
| mmu04024:cAMP
signaling pathway | 30 | 4.9E-4 |
|
mmu04923:Regulation of lipolysis in
adipocytes | 13 | 1.1E-3 |
| mmu04390:Hippo
signaling pathway | 24 | 1.2E-3 |
| mmu04020:Calcium
signaling pathway | 26 | 2.7E-3 |
|
mmu04972:Pancreatic secretion | 17 | 3.8E-3 |
|
mmu00561:Glycerolipid metabolism | 12 | 4.2E-3 |
| mmu04911:Insulin
secretion | 15 | 5.6E-3 |
|
mmu04727:GABAergic synapse | 15 | 6.2E-3 |
|
mmu04512:ECM-receptor interaction | 15 | 6.9E-3 |
|
mmu04261:Adrenergic signaling in
cardiomyocytes | 20 | 2.1E-2 |
| mmu04022:cGMP-PKG
signaling pathway | 22 | 2.2E-2 |
|
mmu04810:Regulation of actin
cytoskeleton | 26 | 2.3E-2 |
|
mmu04726:Serotonergic synapse | 18 | 2.4E-2 |
|
mmu04725:Cholinergic synapse | 16 | 2.6E-2 |
|
mmu04728:Dopaminergic synapse | 18 | 2.8E-2 |
|
mmu04610:Complement and coagulation
cascades | 12 | 3.0E-2 |
| mmu04010:MAPK
signaling pathway | 29 | 3.2E-2 |
| mmu04970:Salivary
secretion | 12 | 3.3E-2 |
| mmu04360:Axon
guidance | 17 | 3.8E-2 |
|
mmu04062:Chemokine signaling pathway | 23 | 4.6E-2 |
PPI network
A total of 419 nodes and 1,343 edges were involved
in the PPI for T1DM, whereas 1,416 nodes and 11,077 edges were
involved in that for T2DM (data not shown). The top 10 proteins in
each of T1- and T2DM with a relatively high degree of connectivity
in the PPI network are presented in Table
V.
 | Table V.Top 10 proteins with relatively high
connectivity degrees in the protein-protein interaction networks
for T1- and T2DM. |
Table V.
Top 10 proteins with relatively high
connectivity degrees in the protein-protein interaction networks
for T1- and T2DM.
| T1DM | T2DM |
|---|
|
|
|---|
| Rank | Protein | Degree | Rank | Protein | Degree |
|---|
|
1 | Lrrk1 | 76 | 1 | Top2a | 171 |
|
2 | Trp53 | 65 | 2 | Cdk1 | 144 |
|
3 | Actb | 60 | 3 | Plk1 | 126 |
|
4 | Actr1b | 42 | 4 | Aurkb | 120 |
|
5 | Fn1 | 40 | 5 | Plk4 | 116 |
|
6 | Yes1 | 38 | 6 | Ccnb1 | 115 |
|
7 | Mapk12 | 38 | 7 | Aurka | 114 |
|
8 | Prkacb | 33 | 8 | Mki67 | 113 |
|
9 | Pten | 32 | 9 | Kif11 | 112 |
| 10 | Ywhaz | 28 | 10 | Cdc20 | 111 |
Extent of enriched function and
topological structure analysis for the modules of the PPI
networks
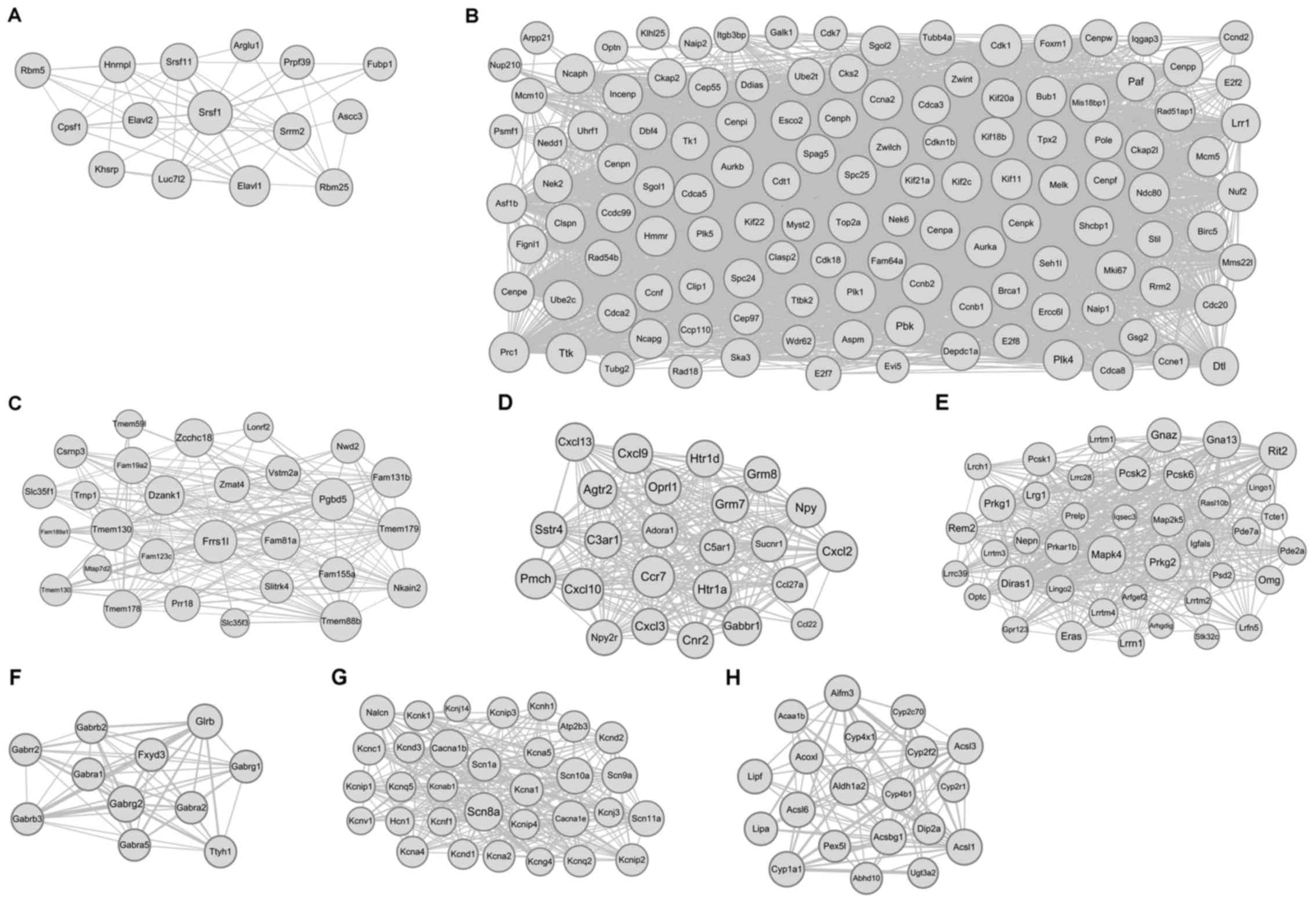
Based on the module analysis by ClusterONE, a single
module was determined in the PPI for T1DM (Fig. 2A) and 7 modules for T2DM (Fig. 2B-H). The information on the modules,
including node number, P-value, protein with highest degree of
connectivity, and numbers of GO terms and KEGG pathways are shown
in Table VI.
 | Table VI.Information on the modules of the
protein-protein interaction networks in T1- and T2DM. |
Table VI.
Information on the modules of the
protein-protein interaction networks in T1- and T2DM.
| Module | Node number | P-value | Protein with
highest connectivity degree (degree) | GO term
number | KEGG pathway
number |
|---|
| T1DM |
| 1 | 15 | 9.4E-5 | Srsf1 (22) | 18 | 0 |
| T2DM |
| 1 | 125 | 4.2E-14 | Top2a (171) | 151 | 8 |
| 2 | 28 | 2.9E-10 | Frrs1l (28) | 2 | 0 |
| 3 | 24 | 8.3E-8 | Ccr7 (46) | 79 | 5 |
| 4 | 41 | 1.0E-4 | Mapk4 (72) | 40 | 4 |
| 5 | 11 | 2.8E-4 | Gabrg2 (25) | 37 | 5 |
| 6 | 32 | 3.4E-4 | Scn8a (52) | 74 | 4 |
| 7 | 20 | 6.0E-4 | Aldh1a2 (30) | 34 | 9 |
Discussion
DPN is among the most frequent complications for T1-
and T2DM. It is generally considered that DPN results from damage
by hyperglycemia, regardless of the type of DM (3). In recent decades, distinct molecular
functions and morphometric abnormalities between DPN in T1- and
T2DM have been reported (7,8). In addition, it has been proposed that
the therapeutic interventions for DPN should differ based on the
type of diabetes (7). For DPN in
T1DM, it has been suggested that improved glycemic control may
preserve nerve function and/or decrease the likelihood of
developing DPN (6). By contrast, for
DPN in T2DM, not only glycemic control, but treatments for other
risk factors including obesity may be equally critical in
preventing DPN (6). Therefore, since
the molecular mechanisms underlying the pathogenesis of DPN in each
type of DM may differ, understanding the mechanisms may lead to
novel therapeutic approaches for prevention or treatment.
In the present study, there were marked differences
between the gene expression profiles, GO terms and KEGG pathways of
DEGs in the sciatic nerves of T1- and T2DM mouse models. This was
in accordance with previous studies revealing considerable
alterations in global gene expression profiles of several tissues
including skeletal muscles and adipose tissue (21), and organs such as the intestine
(22), liver (23) and brain (24) in the two types of DM. It is
established that T1DM is characterized by destruction of pancreatic
islet cells by autoimmunity, with loss of pancreatic insulin
production, while T2DM is a metabolic disease with high pancreatic
insulin production in the setting of insulin resistance (6). Therefore, the different insulin level
and insulin signaling pathways between the two types of DM may
converge on and modulate the transcription of genes (25); it thus seemed reasonable to search for
altered gene expression patterns in DPN between T1- and T2DM models
characterized by absolute or relative insulin deficiency.
A number of DEGs unique to the sciatic nerve of T1DM
mice were associated with cell proliferation, as well as the
mitogen-activated protein kinase signaling pathway, which is a
critical pathway for cell proliferation (26). In neuropathic pain, a correlation has
been determined between the proliferation of nerve cells including
microglia and astrocyte and the abnormal pain responses (27,28). In
addition, previous study has demonstrated that the proliferation of
glia cells including astrocyte was associated with structural
changes in the nervous system, such as axonal growth (29). Furthermore, with loss of C-fibers,
increased frequencies of denervated Schwann cells and regenerating
fibers have been identified in T1DM mice but not in T2DM mice
(8). Therefore, the DEGs associated
with cell proliferation may serve a crucial role during the
pathological changes of DPN in T1DM.
Insulin resistance is among the major factors that
leads to the development and progression of complications in T2DM
(30). Regarding DPN, it has been
suggested that perturbation of insulin receptor signaling due to
insulin resistance may cause neurons to become more vulnerable to
metabolic insults and contribute to the pathogenesis of neuropathy
(31). However, how gene expression
is altered under the insulin-resistant state in nerve tissue
remains unclear for T2DM. In the present study, a number of unique
GO terms and signaling pathways were determined for the sciatic
nerve profile of T2DM mice, which may result from an
insulin-resistant state. For instance, a majority of the GO terms
among the top 20 enriched terms unique to T2DM were related to the
biological processes associated with cell junctions, ion activity
and membrane activity. Previous studies indicated that the
insulin-resistant state induced phosphorylation and downregulated
of the expression of certain connexin (Cx) proteins including Cx43,
which may constitute a potential mechanism underlying the
pathogenesis of insulin resistance and its complications (32,33).
Furthermore, it was demonstrated that Ca2+ overload in
the mitochondria caused production of superoxide and functional
impairment of multiple tissues, which may result in β-cell failure
and insulin resistance in target tissues, further aggravating the
complications of diabetes (34).
Therefore, it may be speculated that the impairment of nerve
tissues under the insulin-resistant state is attributable to these
DEGs in the aforementioned biological processes in T2DM.
In addition, it was apparent that the efficacy of
different treatment strategies for the two types of DM may be
attributed to certain DEGs. For example, treatment of obesity
appeared as critical as glycemic control for preventing the
development of DPN in T2DM (5). In
the present study, C-C chemokine receptor 7 (Ccr7), which was among
the proteins with the highest degrees of connectivity in the PPI
network for T2DM, could interact with the chemokine ligand 19
(Ccl19). The Ccl19-Ccr7 pathway may serve an important role in
development of high-fat-induced obesity and subsequent insulin
resistance (35). Therefore, it may
be suggested that a potential treatment strategy is through Ccr7
targeting to alleviate insulin resistance and neuropathy in
T2DM.
Limitation of the current study included the data
being obtained from online databases, meaning the genetic
backgrounds of the mice could vary substantially. Further genetic
analyses are therefore warranted to identify genes and determine
the molecular differences in neuropathy between the two types of DM
based on different strains of mice.
In conclusion, the present study revealed the gene
expression profiles and signaling pathways associated with the
sciatic nerve in T1- and T2DM mouse models. The DEGs and signaling
pathways may indicate unique biological processes and treatment
strategies for the two types of DM. Further molecular biological
experiments are required to validate the function of the DEGs and
signaling pathways in DPN.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81701104), the
Natural Science Foundation of Guangdong Province, China (grant no.
2016A030310157), the Technology Planning Project of Guangdong
Province, China (grant no. 2016A020220010) and the Technology
Planning Project of Guangzhou, China (grant no. 201604020120).
Availability of data and materials
All data used and/or analyzed during this study are
included in this published article.
Authors contributions
XL designed the study and aided in drafting of the
manuscript. YG and ZLQ performed the bioinformatics analysis and
drafted the manuscript. DZL and GLS analyzed the data. ZQH and YCG
interpreted the results and prepared the figures. XL and ZQH edited
and revised manuscript. All authors approved the final version of
the manuscript to be published.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vincent AM, Callaghan BC, Smith AL and
Feldman EL: Diabetic neuropathy: Cellular mechanisms as therapeutic
targets. Nat Rev Neurol. 7:573–583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tesfaye S and Selvarajah D: Advances in
the epidemiology, pathogenesis and management of diabetic
peripheral neuropathy. Diabetes Metab Res Rev. 28 Suppl 1:8–14.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Premkumar LS and Pabbidi RM: Diabetic
peripheral neuropathy: Role of reactive oxygen and nitrogen
species. Cell Biochem Biophys. 67:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Levitt NS, Stansberry KB, Wynchank S and
Vinik AI: The natural progression of autonomic neuropathy and
autonomic function tests in a cohort of people with IDDM. Diabetes
Care. 19:751–754. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rathmann W, Ziegler D, Jahnke M, Haastert
B and Gries FA: Mortality in diabetic patients with cardiovascular
autonomic neuropathy. Diabet Med. 10:820–824. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
OBrien PD, Sakowski SA and Feldman EL:
Mouse models of diabetic neuropathy. ILAR J. 54:259–272. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Callaghan BC, Hur J and Feldman EL:
Diabetic neuropathy: One disease or two? Curr Opin Neurol.
25:536–541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sima AA and Kamiya H: Diabetic neuropathy
differs in type 1 and type 2 diabetes. Ann NY Acad Sci.
1084:235–249. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kamiya H, Murakawa Y, Zhang W and Sima AA:
Unmyelinated fiber sensory neuropathy differs in type 1 and type 2
diabetes. Diabetes Metab Res Rev. 21:448–458. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stevens MJ, Zhang W, Li F and Sima AA:
C-peptide corrects endoneurial blood flow but not oxidative stress
in type 1 BB/Wor rats. Am J Physiol Endocrinol Metab.
287:E497–E505. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu W, Han C, Li M, Zhang J and Li L:
Revealing the underlying mechanism of diabetic nephropathy viewed
by microarray analysis. Exp Clin Endocrinol Diabetes. 123:353–359.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He K, Lv W, Zhang Q, Wang Y, Tao L and Liu
D: Gene set enrichment analysis of pathways and transcription
factors associated with diabetic retinopathy using a microarray
dataset. Int J Mol Med. 36:103–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lacroix-Fralish ML, Tawfik VL, Tanga FY,
Spratt KF and DeLeo JA: Differential spinal cord gene expression in
rodent models of radicular and neuropathic pain. Anesthesiology.
104:1283–1292. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Qu S, Liang A, Jiang H and Wang
H: Gene expression microarray analysis of the sciatic nerve of mice
with diabetic neuropathy. Int J Mol Med. 35:333–339. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiggin TD, Kretzler M, Pennathur S,
Sullivan KA, Brosius FC and Feldman EL: Rosiglitazone treatment
reduces diabetic neuropathy in streptozotocin-treated DBA/2J mice.
Endocrinology. 149:4928–4937. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pande M, Hur J, Hong Y, Backus C, Hayes
JM, Oh SS, Kretzler M and Feldman EL: Transcriptional profiling of
diabetic neuropathy in the BKS db/db mouse: A model of type 2
diabetes. Diabetes. 60:1981–1989. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Ma SH, Tao R, Xia LJ, Liu L and
Jiang YH: Gene expression profile changes in rat dorsal horn after
sciatic nerve injury. Neurol Res. 39:176–182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang YL, Xiang RL, Yang C, Liu XJ, Shen
WJ, Zuo J, Chang YS and Fang FD: Gene expression profile of human
skeletal muscle and adipose tissue of Chinese Han patients with
type 2 diabetes mellitus. Biomed Environ Sci. 22:359–368.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun J, Wang D and Jin T: Insulin alters
the expression of components of the Wnt signaling pathway including
TCF-4 in the intestinal cells. Biochim Biophys Acta. 1800:344–351.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsumoto K and Yokoyama S: Gene
expression analysis on the liver of cholestyramine-treated type 2
diabetic model mice. Biomed Pharmacother. 64:373–378. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abdul-Rahman O, Sasvari-Szekely M, Ver A,
Rosta K, Szasz BK, Kereszturi E and Keszler G: Altered gene
expression profiles in the hippocampus and prefrontal cortex of
type 2 diabetic rats. BMC Genomics. 13:812012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mounier C and Posner BI: Transcriptional
regulation by insulin: From the receptor to the gene. Can J Physiol
Pharmacol. 84:713–724. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schuelert N, Gorodetskaya N, Just S, Doods
H and Corradini L: Electrophysiological characterization of spinal
neurons in different models of diabetes type 1- and type 2-induced
neuropathy in rats. Neuroscience. 291:146–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Echeverry S, Shi XQ and Zhang J:
Characterization of cell proliferation in rat spinal cord following
peripheral nerve injury and the relationship with neuropathic pain.
Pain. 135:37–47. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun C, Zhang J, Chen L, Liu T, Xu G, Li C,
Yuan W, Xu H and Su Z: IL-17 contributed to the neuropathic pain
following peripheral nerve injury by promoting astrocyte
proliferation and secretion of proinflammatory cytokines. Mol Med
Rep. 15:89–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guzen FP, de Almeida Leme RJ, de Andrade
MS, de Luca BA and Chadi G: Glial cell line-derived neurotrophic
factor added to a sciatic nerve fragment grafted in a spinal cord
gap ameliorates motor impairments in rats and increases local
axonal growth. Restor Neurol Neurosci. 27:1–16. 2009.PubMed/NCBI
|
|
30
|
Chang-Chen KJ, Mullur R and
Bernal-Mizrachi E: Beta-cell failure as a complication of diabetes.
Rev Endocr Metab Disord. 9:329–343. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim B, McLean LL, Philip SS and Feldman
EL: Hyperinsulinemia induces insulin resistance in dorsal root
ganglion neurons. Endocrinology. 152:3638–3647. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bian O, Zhang H, Guan Q, Sun Y and Zeng D:
High-dose insulin inhibits gap junction intercellular communication
in vascular smooth muscle cells. Mol Med Rep. 12:331–336. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takenaka T, Inoue T, Okada H, Ohno Y,
Miyazaki T, Chaston DJ, Hill CE and Suzuki H: Altered gap
junctional communication and renal haemodynamics in Zucker fatty
rat model of type 2 diabetes. Diabetologia. 54:2192–2201. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ly LD, Xu S, Choi SK, Ha CM, Thoudam T,
Cha SK, Wiederkehr A, Wollheim CB, Lee IK and Park KS: Oxidative
stress and calcium dysregulation by palmitate in type 2 diabetes.
Exp Mol Med. 49:e2912017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sano T, Iwashita M, Nagayasu S, Yamashita
A, Shinjo T, Hashikata A, Asano T, Kushiyama A, Ishimaru N,
Takahama Y, et al: Protection from diet-induced obesity and insulin
resistance in mice lacking CCL19-CCR7 signaling. Obesity (Silver
Spring). 23:1460–1471. 2015. View Article : Google Scholar : PubMed/NCBI
|