Introduction
Cervical cancer is the fourth most frequent cancer
amongst women worldwide, resulting in ~570,000 new casesin 2018,
90% of the 311,000 women with cervical cancer died (1). Human papillomavirus (HPV) is the most
common sexually transmitted infection and has been recognized as an
important risk factor for cervical cancer (2,3). There
are >100 types of HPVs, which are categorized into low- and
high-risk HPV (4-7).
High-risk HPVs include HPV16 and HPV18 amongst several others, and
are the causative agents in ≥90% of cervical cancer cases, whilst
also being linked to >50% of other types of anogenital cancer
(7-9).
High-risk HPV (16 and 18)-associated cervical cancer is driven by
two major viral oncoproteins, E6 and E7, which are associated with
the tumor suppressor genes p53 and retinoblastoma 1,
respectively, degrading them (10-15).
HPV16 E6 was found to upregulate DNA methyltransferase I (DNMT1)
expression and suppress p53 in cells (16-19).
Moreover, previous studies have found that E7 displayed an
interaction with DNMT1, which led to aberrant methylation of the
cellular genome, resulting in silencing of tumor suppressor genes
(20-23).
Several groups of researchers have studied the
association between HPV infection and promoter methylation.
Chalertpet et al (24)
reported that HPV16 E7 can induce promoter methylation in
CCNA1. However, Yanatatsaneejit et al (25) in 2020 demonstrated that both HPV E6
and E7 induced promoter methylation in death-associated protein
kinase 1 (DAPK1) and cell adhesion molecule 1
(CADM1), respectively. Moreover, their study indicated that
the E7 protein can bind to the promoter region of CADM1, and
chromatin immunoprecipitation indicated that it may bind to DNMT1
through the same mechanism as CCNA1 (25). Na Rangsee et al (26), using readings from mass spectrometry
followed by STRING database protein network analysis, showed that
E7 possibly formed a complex with a set of transcription factors,
including SP1, which links with the Yin yang 1 (YY1) transcription
factor contributing to the E7 mediated hypermethylation of the
genes. This highlighted the possibility of E7 combining with DNMT1
through a transcription factor to the promoter region of the
CCNA1 gene, resulting in promoter methylation. Thus,
studying other tumor suppressor genes that share the same sequence
as the CCNA1 promoter where HPV16 E7 can bind may highlight
novel potential targets. In order to investigate this possibility,
bioinformatics analysis was performed and the results revealed that
YY1 was found to bind to the promoter region of CCNA1. Here,
other genes which shared the same YY1 binding site as CCNA1
promoter were explored. CGB3 and NOP56 were selected
to investigate the induction of promoter methylation by HPV16
E7.
The CGB3 gene plays critical roles in
increased expression of the CGB subunit in ovarian cancer through
demethylation of the CGB promoter (27) and malignant transformation of
non-trophoblastic cells (28,29).
The NOP56 gene is involved in several oncogenic roles in
more than five types of cancers (30); however, there is no evidence linking
it to cervical cancer, to the best of our knowledge. These findings
lead to the investigation of the role of HPV16 E7 in cervical
cancer progression through promoter methylation of these two
genes.
As methylation does not alter DNA sequences,
previous studies have focused on using inhibitors of DNA
methyltransferases, including 5'-Azacytidine (5-aza), which can
potentially be used as anticancer agents (31-33).
Aberrant DNA methylation is a biochemical process that can be
reversed using demethylating agents (34). 5-aza was the first DNA
methyltransferase inhibitor approved by the U.S. Food and Drug
Administration for use as a chemotherapeutic against
myelodysplastic syndrome, a heterogeneous bone marrow disorder
(35). 5-aza covalently binds to
DNMT leading to a reduction in its DNA methyltransferase activity
(32). This results in the loss of
methylation in specific gene regions and activates the expression
of the associated genes (36-38).
Moreover, several studies have reported that the association
between DNA methylation and repression of gene expression can lead
to cancer progression (39-41).
Therefore, re-expression of methylated genes in cervical cancer
cell lines following treatment with a demethylating agent, such as
5-aza, should be further investigated to assess the effect of
inhibiting the methylation rate and whether it can increase the
expression of tumor suppressor genes (31,42).
The present study investigated whether HPV16 E7
could induce promoter methylation and decrease the expression of
CGB3 and NOP56. The study further assessed the
effects of 5-aza treatment on promoter methylation and gene
expression in cervical cancer C33A and SiHa cell lines to
demonstrate the association between gene methylation and the
regulation of tumor gene expression. This study may serve as an
alternative strategy of drug therapy for patients with cervical
cancer with aberrant gene promoter methylation (42).
Materials and methods
Bioinformatics analysis
PROMO version 8.3 from the TRANSFAC database
(alggen.lsi.upc.es) was used to select the genes
of interest containing YY1 binding sites with CCNA1
(43,44). JASPAR database (jaspar.genereg.net) was used to separately analyze the
YY1 binding site sequences in the selected genes with reference to
the CCNA1 gene (45).
Cell lines and culture
Human cervical carcinoma cell lines [SiHa (HPV type
16) and C33A (HPV-)] were purchased from the American Type Culture
Collection. Cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% antibiotic-antimycotic (Gibco; Thermo
Fisher Scientific, Inc.) at 37˚C with 5% CO2 in a
humidified incubator.
Transfection
For HPV 16 E7 overexpression, 3x105
cells/ml C33A cells were seeded into a 6-well plate and incubated
overnight at 37˚C. Subsequently, cells were transfected with 2 µg
HPV 16 E7 plasmid (E7; Invitrogen; Thermo Fisher Scientific, Inc.)
and pcDNA 3.1/myc-HIS empty vector (PC; Invitrogen; Thermo Fisher
Scientific, Inc.) using TurboFect reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A 72 h
post-transfection, DNA and RNA were extracted from each sample to
assess promoter methylation and gene expression, respectively,
using specific primers for each gene.
5-aza treatment
To evaluate gene methylation and expression, cells
were treated with 5-aza (Sigma-Aldrich; Merck KGaA). Briefly, SiHa
and C33A cells (3x105 cells/ml) were seeded 1 day before
5-aza treatment. The following day fresh DMEM containing 5-aza
(SiHa, 0, 20, 30 and 40 µM; C33A, 0, 3,5 and 7 µM) was added to the
cells for 5 consecutive days, being replaced every 24 h until
analysis (24).
Isolation of DNA
DNA was extracted from SiHa and C33A cells using 10%
SDS (Sigma-Aldrich; Merck KGaA), lysis buffer II (0.75 M NaCl and
0.024 M EDTA at pH 8) and 20 mg/ml proteinase K (Invitrogen; Thermo
Fisher Scientific, Inc.), digested by incubating at 50˚C overnight.
Subsequently, phenol/chloroform extraction and 100% ethanol
precipitation were performed. DNA was air-dried and resuspended in
dH2O (46). DNA
concentration was determined using a NanoDrop 2000c
spectrophotometer (Thermo Fisher Scientific, Inc.).
Sodium bisulfite treatment and
methylation-specific PCR (MSP)
DNA (750 ng) for each sample was subjected to
bisulfite treatment using the EZ DNA Methylation-Gold kit (Zymo
Research Corp.) according to the manufacturer's protocol. Eluted
DNA was used to perform MSP using methylated and unmethylated
specific primers (Table I). The
annealing temperature for CGB3 was 54˚C and for NOP56 it was
52˚C. The thermocycling conditions were 95˚C for 15 min; followed
by 27 cycles of 95˚C for 45 sec, the respective annealing
temperature for 45 sec and 72˚C for 45 sec; with a final extension
step of 72˚C for 7 min. Subsequently, 10 µl PCR product was
observed by gel electrophoresis using an 8% acrylamide gel and
stained with SYBR reagent (Lonza Group, Ltd.). The methylated and
unmethylated band intensities of each sample were visualized and
measured using a Storm 840 (Amersham Biosciences) and ImageJ
version 2 (National Institute of Health). The EpiTect control DNA
set (Qiagen GmbH) contained positive controls for methylation and
unmethylation. The experiments were performed in triplicate.
 | Table IPrimer sequences, amplicon sizes,
annealing temperature and conditions for methylation-specific
PCR. |
Table I
Primer sequences, amplicon sizes,
annealing temperature and conditions for methylation-specific
PCR.
| Gene | Sequence,
5'-3' | Product size,
bp | Annealing
temperature, ˚C | Number of
cycles |
|---|
| CGB3 | | | | |
|
Methylated
forward |
CGGGTTGAATTTTTCGTTGGC | 116 | 52 | 27 |
|
Methylated
reverse |
CCCAAAAAAAACGCGACTTCG | | | |
|
Unmethylated
forward |
GGGTTTGGGTTGAATTTTTTGTTGGT | 127 | 55 | 27 |
|
Unmethylated
reverse |
CAACCTCCCAAAAAAAACACAACTTCA | | | |
| NOP56 primer
I | | | | |
|
Methylated
forward |
TATTTTTTATTATATTTTGGAATC | 64 | 38 | 27 |
|
Methylated
reverse |
ATTAAATTATTTTAACCGTCG | | | |
|
Unmethylated
forward |
GTATTTTTTATTATATTTTGGAATT | 68 | 42 | 27 |
|
Unmethylated
reverse |
AATATTAAATTATTTTAACCATCA | | | |
| NOP56 primer
II | | | | |
|
Methylated
forward |
CGTTTATTTTGATGTATTTACGAC | 120 | 50 | 27 |
|
Methylated
reverse |
ATATCTACTTACGAATCAAAATCG | | | |
|
Unmethylated
forward |
TGGAATTGTTTATTTTGATGTATTTATGATG | 132 | 52 | 27 |
|
Unmethylated
reverse |
AATATTAAATTATTTTAACCATCA | | | |
Preparation of RNA and cDNA
synthesis
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Subsequently, total RNA (1 µg) from each sample was reverse
transcribed into cDNA using the RevertAid first-strand cDNA
synthesis kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
PCR
PCR was performed to assess the expression of each
gene in SiHa and C33A cells using specific forward and reverse
primers for CGB3 expression in HPV 16 E7 transfected cell.
The PCR mixture contained 10X PCR buffer (Qiagen GmbH), 0.2 mM
dNTPs (New England BioLabs, Inc.), 0.3 µM forward and reverse
primer (Table I) and 1 U HotStartaq
DNA polymerase (Qiagen GmbH). GAPDH was used as an internal
control (Table II). PCR products
(10 µl) were subjected to electrophoresis on an 8% acrylamide gel
and stained with SYBR. The experiment was performed in
triplicate.
 | Table IIPrimer sequences, amplicon sizes,
annealing temperatures and conditions for PCR. |
Table II
Primer sequences, amplicon sizes,
annealing temperatures and conditions for PCR.
| Gene | Sequence,
5'-3' | Product size,
bp | Annealing
temperature, ˚C | Number of
cycles |
|---|
| Human papilloma
virus 16 E7 | | | | |
|
Forward |
GGGCAATTAAATGACAGCTCAG | 142 | 56 | 30 |
|
Reverse |
GTGTGCTTTGTACGCACAACC | | | |
| GAPDH | | | | |
|
Forward |
CAGCCGCATCTTCTTTTG | 96 | 56 | 28 |
|
Reverse |
GCCCAATACGACCAAATC | | | |
| GAPDH1 | | | | |
|
Forward |
TGGAAGGACTCATGACCACAG | 163 | 56 | 28 |
|
Reverse |
TTCAGCTCAGGGATGACCTT | | | |
| Chorionic
gonadotropin subunit 3 | | | | |
|
Forward |
CAACACCACCATCTGTGC | 194 | 56 | 28 |
|
Reverse |
GGCAGAGTGCACATTGAC | | | |
| Nucleolar protein
56 | | | | |
|
Forward |
CAGCATCGTTCGTCTGGTGG | 105 | 56 | 28 |
|
Reverse |
AGGCGGAGGTCCTCATGAAC | | | |
Quantitative PCR (qPCR)
CGB3 and NOP56 mRNA expression in
5-aza-treated, and HPV 16 E7 and NOP56 gene expression
levels in HPV 16 E7 transfected cells were determined by qPCR using
a 7500-fast qPCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and Power SYBR-Green PCR MasterMix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The qPCR was performed
using specific primers (Table II),
with GAPDH as the reference gene. The thermocycling
condition were: 95˚C for 10 min; followed by 40 cycles of 95˚C for
15 sec, 56˚C for 30 sec and 72˚C for 45 sec. The mRNA expression
levels were quantified using the 2-IICq method (47).
Statistical analysis
To assess gene promoter methylation and expression
levels in 5-aza-treated cells, a one-way ANOVA followed by a
Dunnett's post hoc test was performed using GraphPad Prism version
5 (GraphPad Software, Inc.). P≤0.05 was considered to indicate a
statistically significant difference. Student's t-tests (unpaired)
was used to compare CGB3 and NOP56 expression and
methylation in SiHa and C33A cells, respectively, as well as
expression and methylation of CGB3 and NOP56 in the
HPV 16 E7- and PC-transfected cells.
Results
Bioinformatics analysis
YY1 was found to be the binding to the promoter of
CCNA1, CGB3 and NOP56 based on PROMO analysis
(Fig. 1). Moreover, the results
from JASPAR demonstrated that the binding sequence of YY1 in the
CCNA1 gene shared 80% similarity in sequence, GCCATG and
TCCATG (Fig. 2A), to that present
in CGB3 GCCATC denoted by * in (Fig. 2B) and in NOP56 TCCATC denoted
by ** in (Fig. 2C), respectively.
Thus, both genes were selected for further analysis.
Gene expression and methylation in
HPV+/- cervical cancer cell lines
To assess the expression of CGB3 and
NOP56 genes, PCR was performed in SiHa (HPV+) and
C33A (HPV-) cancer cell lines. The results showed that
both CGB3 and NOP56 exhibited lower expression in
SiHa cells (26.74 and 38.42%, respectively) compared with that
observed in C33A cells (73.26 and 44.5%), respectively (Fig. 3A and C). GAPDH was used for CGB3 gene and
GAPDH1 primer was used for NOP56 gene normalization in SiHa
and C33A cells. The bar graphs shows the mean ± standard error of
the mean of CGB3 (P=0.001; Fig.
3B) and NOP56 expression (P=0.004; Fig. 3D).
Furthermore, methylation status was also observed
for both the genes, and compared between SiHa and C33A cells. The
results demonstrated that the methylation of CGB3 and
NOP56 (primer II) in SiHa cells was 61.18 and 53.94%
compared with C33A cells (38.81 and 46.05%), respectively (Fig. 4A and C). The bar graph shows the mean ± standard
error of the mean of CGB3 (P=0.004; Fig. 3B) and NOP56 methylation
(P=0.045; Fig. 3D). The results
indicated that the presence of HPV may influence the expression and
methylation of CGB3 and NOP56 genes.
HPV 16 E7 induces promoter methylation
and decreases gene expression
To assess whether HPV E7 induced gene promoter
methylation and thus decreased expression, HPV 16 E7 was
overexpressed in C33A (HPV-) cells. The results
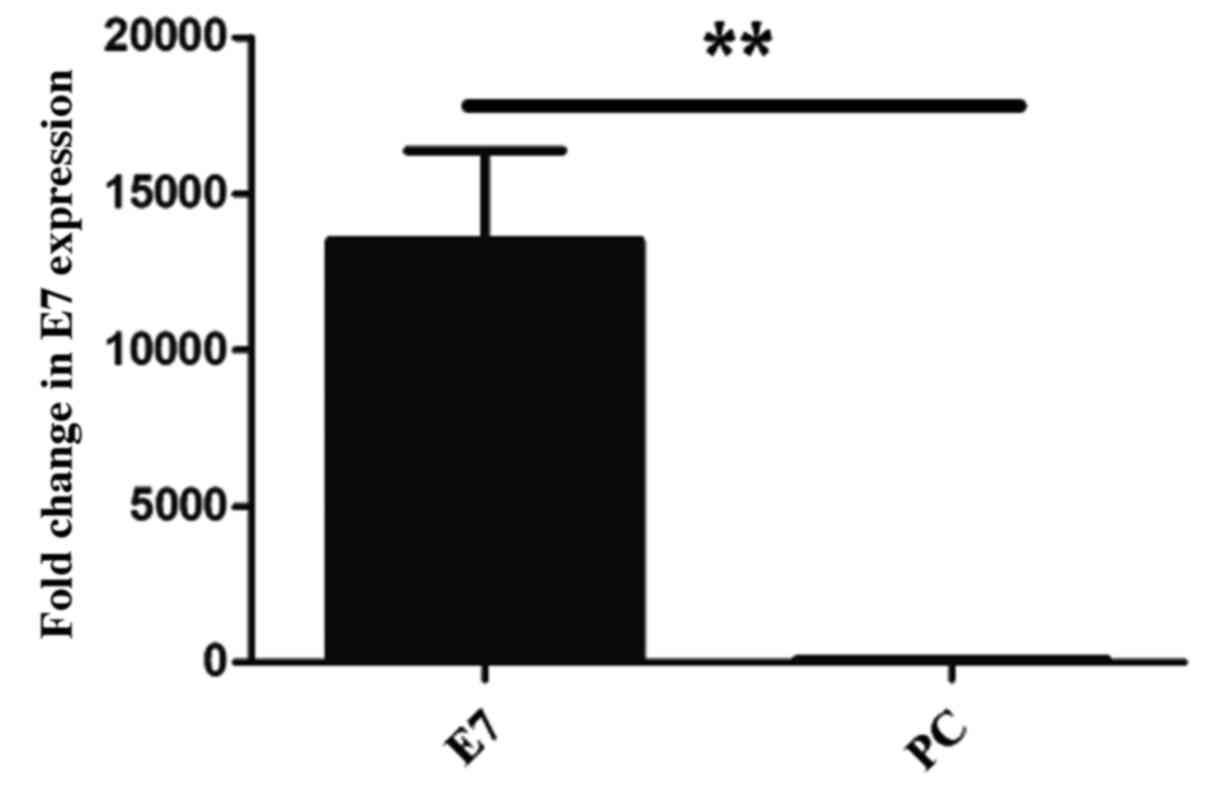
demonstrated that the expression of HPV 16 E7 was significantly
higher in HPV 16 E7 transfected cells (P=0.001) compared with the
PC transfected cells (Fig. 5).
Furthermore, alterations to gene promoter
methylation and expression were assessed in HPV 16 E7 transfected
C33A cells. At 72 h post-transfection, the band intensities of
CGB3 and NOP56 were compared between E7- and
PC-transfected cells. To measure gene expression and methylation,
reverse transcription-PCR and MS-PCR were performed, respectively.
The gene expression levels of CGB3 were significantly
reduced (P=0.02) in HPV 16 E7 transfected cells (band intensity,
37.2%) compared with that in the PC transfected cells (band
intensity, 62.7%) (Fig. 6A). The
bar graph shows the mean ± standard error of CGB3 expression
(Fig. 6B). NOP56 expression
level in HPV 16 E7 transfected C33A cells was significantly
decreased (P<0.0001) compared with that in the PC transfected
cells (Fig. 7). GAPDH was
used as an internal control. The expression of NOP56 was
measured by qPCR.
Promoter methylation of both genes was significantly
increased in HPV 16 E7 overexpressing C33A cells compared with that
in the PC transfected cells. In HPV 16 E7 transfected C33A cells,
the band intensities of CGB3 and NOP56 (Primer I)
were 54.21 and 55.01%, respectively, which were significantly
higher than those in the PC transfected cells (39.36 and 41.54%;
P=0.005 and P=0.0004, respectively). The methylation positive
control displayed band intensities of 6.43 and 3.45%, respectively
(Fig. 8A and C). The bar graph shows the mean ± standard
error of the mean of both CGB3 and NOP56 expression
(Fig. 8B and D), respectively. The unmethylated forms of
CGB3 and NOP56 (Primer I) in HPV 16 E7 transfected
cells gradually decreased (data not shown).
Effect of 5-aza on gene promoter
methylation and gene expression of CGB3 and NOP56
An association between promoter methylation and gene
expression was confirmed by measuring mRNA expression levels in
SiHa and C33A cells following 5-aza treatment. The C33A and SiHa
cancer cells were treated continuously for 5 days with 5-aza at
concentrations of 0, 3, 5 and 7 or 0, 20, 30 and 40 µM,
respectively. Subsequently, DNA and RNA were extracted to measure
promoter methylation and expression levels of each gene in cells
following treatment with different concentrations of 5-aza.
The CGB3 and NOP56 (Primer II)
promoter methylation results demonstrated that the methylation
status of both gene promoters in C33A and SiHa cells were
significantly decreased following treatment with different
concentrations of 5-aza. The band densities of CGB3 and
NOP56 methylation in C33A cells gradually decreased as the
5-aza concentration increased (0-7 µM; Fig. 9A and C), whereas the unmethylated forms of both
genes in C33A cells increased with increasing 5-aza concentrations
(data not shown). In parallel, the band densities of CGB3
and NOP56 methylation (Primer II) in SiHa cells decreased
with increasing 5-aza concentrations (0-40 µM; Fig. 9B and D). The decrease in the methylation status
of both CGB3 and NOP56 following treatment with
different concentrations of 5-aza in C33A (P<0.0001 and
P=0.0002; Fig. 10A and C) and SiHa (P<0.0001 and P=0.0002;
Fig. 10B and D) cells was significant.
 | Figure 9Treatment of C33A and SiHa cells with
0, 3, 5 and 7 µM or 0, 20, 30 and 40 µM 5-Azacytidine,
respectively. Methylation status of (A) CGB3 in C33A cells,
(B) CGB3 in SiHa cells, (C) NOP56 in C33A cells and
(D) NOP56 in SiHa cells. Neg, distilled water negative
control; CCNA1, cyclin-A1; CGB3, chorionic
gonadotropin subunit 3; NOP56, nucleolar protein 56. |
Gene expression was shown by using reverse
transcription-PCR and qPCR. The band densities of CGB3 and
NOP56 expression in C33A cells gradually increased with
increasing 5-aza concentrations (0-7 µM; Fig. 11A and C). The expression of CGB3 and
NOP56, as determined using qPCR, in C33A cells also
significantly increased with all concentrations of 5-aza treatment
(both P<0.0001; Fig. 11B and
D). Similarly, the band densities
of CGB3 and NOP56 observed in SiHa by RT-PCR
gradually increased with increasing 5-aza concentration (0-40 µM;
Fig. 12A and C). Expression of CGB3 and NOP56 measured
by qPCR in SiHa cells were significantly higher when treated with
40 µM 5-aza (both P<0.0001; Fig.
12B and D).
Discussion
HPV is the primary cause of cervical cancer. There
are several studies showing that HPV 16 E7 is involved in promoter
methylation (22,24-25).
Chalertpet et al (24)
showed that HPV 16 E7 can interact with DNMT1 on the CCNA1
promoter leading to its methylation. Moreover, a study by
Yanatatsaneejit et al (25)
showed that HPV 16 E7 could induce CADM1 promoter
methylation through the same mechanism as CCNA1. In
contrast, DAPK1 promoter methylation could no be induced by
HPV 16 E7(25). Therefore, it was
hypothesized that there would be genes other than CCNA1 and
CADM1 whose promoters are methylated by HPV 16 E7, and that
there would be specific transcription factors binding with HPV 16
E7 on the gene promoter. A study by Na Rangsee et al,used
STRING database protein network analysis, showed that E7 possibly
forms a complex with the set of transcription factors containing a
YY1 domain (26). In the present
study, the binding site of HPV 16 E7 on the CCNA1 promoter
was analyzed to identify the putative transcription factors.
According to bioinformatics analysis, a list of genes was analyzed
based on the presence of the promoter sequence found in the
CCNA1 gene where HPV 16 E7 binds. Interestingly the genes
CGB3 and NOP56 showed positive results. Thus, in
addition to CCNA1 (24) and
CADM1 (25), several other
tumor suppressor genes may be affected by HPV 16 E7 through the
same mechanism as that observed with CCNA1 and CADM1.
To assess this, CGB3 and NOP56 methylation and
expression were observed in two cervical cancer cell lines, HPV
16-positive SiHa and HPV-negative C33A cells. Both CGB3 and
NOP56 expression in SiHa cells had lower band intensities
compared with the C33A cells, suggesting that the presence of HPV
in SiHa may induce promoter methylation and decrease the expression
of the genes. Moreover, the expression of CGB3 and
NOP56 in cells transfected with HPV 16 E7 and PC plasmids to
assess the effect of HPV 16 E7 on the expression of these genes.
Both genes exhibited lower expression in HPV 16 E7 transfected
cells compared with the PC transfected cells, which demonstrated
that HPV16 E7 induced promoter methylation of CGB3 and
NOP56, resulting in the downregulation of these genes. Thus,
the expression changes in CGB3 and NOP56 following
methylation inhibition using the demethylating agent 5-aza was next
assessed.
5-aza interferes with DNA methylation and can be
used to reactivate silenced genes in cancer (31-34).
Examples of genes reactivated following 5-aza treatment include
CD44, GSTP-1, BRCA1, MDR1, MUC2
and GPC3, which possess hypermethylated promoter CpG islands
in tumors (31). Wong et al
(48) showed that
methylation-mediated silencing of gene expression can be reversed
by treatment with the demethylating agent 5-aza. Reactivation of
methylation-silenced genes indicates that the gene is functional,
and that DNA methylation regulates its transcription (39,40).
With regard to the present study, it was necessary to treat SiHa
cells with higher concentrations of 5-aza compared with the C33A
cells, as SiHa is a HPV positive cancer cell line, meaning that
promoter methylation was more prominent in them. Therefore, the
lower concentration of 5-aza may not overcome HPV-induced
methylation in SiHa cells. Following treatment with 5-aza at higher
concentrations of 20, 30 and 40 µM in SiHa cells, and at lower
concentrations of 3, 5, and 7 µM in C33A cells for 5 days, it was
found that the methylation levels of CGB3 and NOP56
were decreased leading to increased expression of both genes. At 40
µM, the re-expression of CGB3 and NOP56 genes was
observed.
In conclusion, the results of the present study
suggest that patients with cervical cancer with HPV infection
should be treated with a higher concentration of 5-aza than
patients without HPV infection (42,49).
Taken together, the findings of the present study showed that 5-aza
may serve as an alternative strategy of drug therapy for patients
with cervical cancer with aberrant gene promoter methylation.
Although the present study demonstrated that HPV16 E7 decreased the
expression of CGB3 and NOP56, and that 5-aza
treatment increased the expression of these genes, it cannot be
concluded that these genes are tumor suppressor genes in cervical
cancer. Further study of the function and effects of these genes in
cervical cancer is thus required.
Acknowledgements
We are grateful to Professor Apiwat Mutirangura,
Center of Excellence in Molecular Genetics of Cancer and Human
Diseases, Chulalongkorn University for providing the laboratory
equipment and valuable suggestions.
Funding
The present study was financially supported by the Thailand
Research Fund and Chulalongkorn University (grant no. RSA5880065),
The National Science and Technology Development Agency, Thailand
(grant no. P-15-50270) and the Second Century Fund (C2F),
Chulalongkorn University.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
PSi performed the experiments, analyzed the data
and wrote the manuscript. KC, JS and NW performed the experiments
and analyzed the data. PSu performed the experiments. PY wrote the
proposal for grants, designed the experiments, analyzed the data
and wrote the manuscript. All authors read and approved the final
manuscript. PSi and KC confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Pineros M, Znaor A, Soerjomataram I and Bray F: Global Cancer
Observatory: Cancer Today. International Agency for Research on
Cancer, Lyon, 2018.
|
|
2
|
Bosch FX, Burchell AN, Schiffman M,
Giuliano AR, de Sanjose S, Bruni L, Tortolero-Luna G, Kjaer SK and
Muñoz N: Epidemiology and natural history of human papillomavirus
infections and type-specific implications in cervical neoplasia.
Vaccine. 26 (Suppl 10):K1–K16. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
zur Hausen H: Papillomaviruses in the
causation of human cancers - a brief historical account. Virology.
384:260–265. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Clifford GM, Smith JS, Plummer M, Muñoz N
and Franceschi S: Human papillomavirus types in invasive cervical
cancer worldwide: A meta-analysis. Br J Cancer. 88:63–73.
2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Burd EM: Human papillomavirus and cervical
cancer. Clin Microbiol Rev. 16:1–17. 2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Egawa N and Doorbar J: The low-risk
papillomaviruses. Virus Res. 231:119–127. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group. Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tulay P and Serakinci N: The role of human
papillomaviruses in cancer progression. J Cancer Metastasis Treat.
2:201–213. 2016.
|
|
9
|
zur Hausen H and de Villiers EM: Human
papillomaviruses. Annu Rev Microbiol. 48:427–447. 1994.
|
|
10
|
Havre PA, Yuan J, Hedrick L, Cho KR and
Glazer PM: p53 inactivation by HPV16 E6 results in increased
mutagenesis in human cells. Cancer Res. 55:4420–4424.
1995.PubMed/NCBI
|
|
11
|
Pim D, Massimi P and Banks L:
Alternatively spliced HPV-18 E6* protein inhibits E6 mediated
degradation of p53 and suppresses transformed cell growth.
Oncogene. 15:257–264. 1997.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Caldeira S, Dong W and Tommasino M:
Analysis of E7/Rb associations. Methods Mol Med. 119:363–379.
2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu EW, Clemens KE, Heck DV and Münger K:
The human papillomavirus E7 oncoprotein and the cellular
transcription factor E2F bind to separate sites on the
retinoblastoma tumor suppressor protein. J Virol. 67:2402–2407.
1993.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Collins AS, Nakahara T, Do A and Lambert
PF: Interactions with pocket proteins contribute to the role of
human papillomavirus type 16 E7 in the papillomavirus life cycle. J
Virol. 79:14769–14780. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Slebos RJ, Lee MH, Plunkett BS, Kessis TD,
Williams BO, Jacks T, Hedrick L, Kastan MB and Cho KR:
p53-dependent G1 arrest involves pRB-related proteins and is
disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl
Acad Sci USA. 91:5320–5324. 1994.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Scheffner M, Werness BA, Huibregtse JM,
Levine AJ and Howley PM: The E6 oncoprotein encoded by human
papillomavirus types 16 and 18 promotes the degradation of p53.
Cell. 63:1129–1136. 1990.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Villa LL: Human papillomaviruses and
cervical cancer. Adv Cancer Res. 71:321–341. 1997.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Schwarz E, Freese UK, Gissmann L, Mayer W,
Roggenbuck B, Stremlau A and zur Hausen H: Structure and
transcription of human papillomavirus sequences in cervical
carcinoma cells. Nature. 314:111–114. 1985.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Au Yeung CL, Tsang WP, Tsang TY, Co NN,
Yau PL and Kwok TT: HPV-16 E6 upregulation of DNMT1 through
repression of tumor suppressor p53. Oncol Rep. 24:1599–1604.
2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Burgers WA, Blanchon L, Pradhan S, de
Launoit Y, Kouzarides T and Fuks F: Viral oncoproteins target the
DNA methyltransferases. Oncogene. 26:1650–1655. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sen P, Ganguly P and Ganguly N: Modulation
of DNA methylation by human papillomavirus E6 and E7 oncoproteins
in cervical cancer. Oncol Lett. 15:11–22. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li L, Xu C, Long J, Shen D, Zhou W, Zhou
Q, Yang J and Jiang M: E6 and E7 gene silencing results in
decreased methylation of tumor suppressor genes and induces
phenotype transformation of human cervical carcinoma cell lines.
Oncotarget. 6:23930–23943. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chalertpet K, Pakdeechaidan W, Patel V,
Mutirangura A and Yanatatsaneejit P: Human papillomavirus type 16
E7 oncoprotein mediates CCNA1 promoter methylation. Cancer Sci.
106:1333–1340. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yanatatsaneejit P, Chalertpet K,
Sukbhattee J, Nuchcharoen I, Phumcharoen P and Mutirangura A:
Promoter methylation of tumor suppressor genes induced by human
papillomavirus in cervical cancer. Oncol Lett. 20:955–961.
2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Na Rangsee N, Yanatatsaneejit P, Pisitkun
T, Somparn P, Jintaridth P and Topanurak S: Host proteome linked to
HPV E7-mediated specific gene hypermethylation in cancer pathways.
Infect Agent Cancer. 15(7)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Śliwa A, Kubiczak M, Szczerba A, Walkowiak
G, Nowak-Markwitz E, Burczyńska B, Butler S, Iles R, Białas P and
Jankowska A: Regulation of human chorionic gonadotropin beta
subunit expression in ovarian cancer. BMC Cancer.
19(746)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bellet D, Lazar V, Bièche I, Paradis V,
Giovangrandi Y, Paterlini P, Lidereau R, Bedossa P, Bidart JM and
Vidaud M: Malignant transformation of nontrophoblastic cells is
associated with the expression of chorionic gonadotropin beta genes
normally transcribed in trophoblastic cells. Cancer Res.
57:516–523. 1997.PubMed/NCBI
|
|
29
|
Sohr S and Engeland K: The tumor
suppressor p53 induces expression of the pregnancy-supporting human
chorionic gonadotropin (hCG) CGB7 gene. Cell Cycle. 10:3758–3767.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gong J, Li Y, Liu CJ, Xiang Y, Li C, Ye Y,
Zhang Z, Hawke DH, Park PK, Diao L, et al: A Pan-cancer analysis of
the expression and clinical relevance of small nucleolar RNAs in
human cancer. Cell Rep. 21:1968–1981. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Christman JK: 5-Azacytidine and
5-aza-2'-deoxycytidine as inhibitors of DNA methylation:
Mechanistic studies and their implications for cancer therapy.
Oncogene. 21:5483–5495. 2002.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Stresemann C and Lyko F: Modes of action
of the DNA methyltransferase inhibitors azacytidine and decitabine.
Int J Cancer. 123:8–13. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Glover AB and Leyland-Jones B:
Biochemistry of azacitidine: A review. Cancer Treat Rep.
71:959–964. 1987.PubMed/NCBI
|
|
34
|
Miranda TB and Jones PA: DNA methylation:
The nuts and bolts of repression. J Cell Physiol. 213:384–390.
2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Müller A and Florek M:
5-Azacytidine/Azacitidine. Recent Results Cancer Res. 184:159–170.
2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lande-Diner L, Zhang J, Ben-Porath I,
Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G and
Cedar H: Role of DNA methylation in stable gene repression. J Biol
Chem. 282:12194–12200. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cheng JC, Matsen CB, Gonzales FA, Ye W,
Greer S, Marquez VE, Jones PA and Selker EU: Inhibition of DNA
methylation and reactivation of silenced genes by zebularine. J
Natl Cancer Inst. 95:399–409. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ferguson AT, Lapidus RG, Baylin SB and
Davidson NE: Demethylation of the estrogen receptor gene in
estrogen receptor-negative breast cancer cells can reactivate
estrogen receptor gene expression. Cancer Res. 55:2279–2283.
1995.PubMed/NCBI
|
|
39
|
Karpf AR and Jones DA: Reactivating the
expression of methylation silenced genes in human cancer. Oncogene.
21:5496–5503. 2002.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2 (Suppl 1):S4–S11.
2005.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sova P, Feng Q, Geiss G, Wood T, Strauss
R, Rudolf V, Lieber A and Kiviat N: Discovery of novel methylation
biomarkers in cervical carcinoma by global demethylation and
microarray analysis. Cancer Epidemiol Biomarkers Prev. 15:114–123.
2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Biktasova A, Hajek M, Sewell A, Gary C,
Bellinger G, Deshpande HA, Bhatia A, Burtness B, Judson B, Mehra S,
et al: Demethylation therapy as a targeted treatment for human
papillomavirus-associated head and neck cancer. Clin Cancer Res.
23:7276–7287. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Messeguer X, Escudero R, Farré D, Núñez O,
Martínez J and Albà MM: PROMO: Detection of known transcription
regulatory elements using species-tailored searches.
Bioinformatics. 18:333–334. 2002.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Farré D, Roset R, Huerta M, Adsuara JE,
Roselló L, Albà MM and Messeguer X: Identification of patterns in
biological sequences at the ALGGEN server: PROMO and MALGEN.
Nucleic Acids Res. 31:3651–3653. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fornes O, Castro-Mondragon JA, Khan A, van
der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M,
Baranašić D, et al: JASPAR 2020: Update of the open-access database
of transcription factor binding profiles. Nucleic Acids Res.
48:D87–D92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Maniatis T, Fritsch EF and Sambrook J:
Isolation of DNA from mammalian cells. In: Molecular Cloning. A
Laboratory Manual. Nolan C (ed). Cold Spring Harbor, New York, NY,
9.16-9.23, 1982.
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wong J, Sia YY, Misso NL, Aggarwal S, Ng A
and Bhoola KD: Effects of the demethylating agent, 5-azacytidine,
on expression of the kallikrein-kinin genes in carcinoma cells of
the lung and pleura. Patholog Res Int. 2011(167046)2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Abdulhaq H and Rossetti JM: The role of
azacitidine in the treatment of myelodysplastic syndromes. Expert
Opin Investig Drugs. 16:1967–1975. 2007.PubMed/NCBI View Article : Google Scholar
|