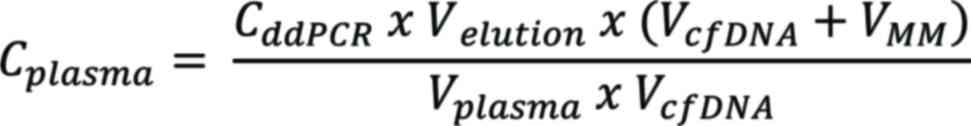Introduction
Improved surgical techniques and the development of
new immunosuppressive drugs have significantly extended the
survival of heart transplant (HTx) recipients. Acute rejection,
which occurs in 25-32% of patients after HTx, still represents a
mortality risk during the first 1-3 years following cardiac
transplantation (1). The current
standard for graft rejection surveillance is endomyocardial biopsy
(EMB), an invasive procedure with rare but potentially serious
complications (1). Therefore, the
development of a new non-invasive method is required.
DNA is located mostly in the nuclei of cells, but at
low concentrations, cell-free (cf)DNA is present in the plasma of
all individuals (2). Donor-derived
cfDNA (ddcfDNA) released into plasma from transplanted organ
necrotic or apoptotic cells may serve as a non-invasive biomarker
for the early detection of allograft rejection (3-5).
This has been suggested for decades, but detection of a graft's DNA
at the recipient's background has thus far proved challenging in
terms of sufficient specificity and throughput. Different methods
have been proposed [shotgun sequencing, target sequencing, digital
droplet PCR (ddPCR)…] to quantify levels of ddcfDNA in recipient
plasma using the detection of donor-specific genotypes (3,6,7).
Changes in recipient cfDNA (e.g., due to leukopenia, leukocytosis
and inflammatory illness) can affect the results of ddcfDNA
fractional determination (%ddcfDNA), leading to falsely elevated or
decreased results (8). This
limitation may be overcome using absolute ddcfDNA quantification.
The combination of fractional and absolute determination including
total cfDNA is recommended for meaningful interpretation of the
results. Fractional determination is possible with all mentioned
methods but absolute quantification has only been validated for
ddPCR (8-10).
Circulating ddcfDNA has been investigated in studies of liver,
kidney and heart transplants (2,3,5,11).
It has been shown that immediately after engraftment, graft cfDNA
reaches high values (>5% of total cfDNA). Within the first 2
weeks after transplantation, ddcfDNA typically exponentially
declines to baseline levels (9).
This may be used to discriminate graft injury. An abnormal
non-exponential decline of ddcfDNA has been observed in certain
patients due to urinary tract infections, hemodynamic problems or
surgical complications (12).
Episodes of rejection in heart and kidney transplants are
accompanied by a significant increase of graft cfDNA (>5-fold)
levels in patients without complications, occurring earlier than
clinical or biochemical markers of rejection (4).
Patients and methods
Patient characteristics
The selection criteria in the present case report
study were noncomplicated heart transplantation (HTx) and
postoperative course with later EMB confirming acute cellular
rejection (ACR) or antibody-mediated rejection (AMR) at the time of
blood collection. The patients were subjected to standard clinical
practice. The timeline for performing EMBs is presented in Table I. Patients who failed to appear at
their scheduled blood draw appointment were not included in the
study. Histology, immunohistochemistry and EMB were performed
according standard protocols and guidelines (13-15).
 | Table ITimeline for performing EMBs,
routinely performed within the first year at our institute. |
Table I
Timeline for performing EMBs,
routinely performed within the first year at our institute.
| EMB | Time after HTx | Frequency |
|---|
| 1st | Between 7th and 10th
days | First EMB is
performed on the 7th day |
| 2nd | On the 21st day | at the earliest;
then, they follow in the |
| 3rd | On the 30th day | frequency of once
every 10 days |
| 4th and 5th | Within 2 months | Once every 14
days |
| 6th | At the end of the 3rd
month | Once a month |
| 7th | At the end of the 6th
month | 3 months apart |
| 8th | 1 year | 6 months apart |
Patient 1 (male; age, 63 years) underwent orthotopic
HTx (OHT) in March 2016 at the Cardio Center of the Institute for
Clinical and Experimental Medicine (Prague, Czech Republic) due to
dilated cardiomyopathy. Standard immunosuppression was applied. The
patient received induction with rabbit antithymocyte globulin
(rATG; Fresenius). The maintenance therapy consisted of tacrolimus
(Astellas Pharma Europe B.V., Leiderdorp), mycophenolate mofetil
(Salutas Pharma) and corticosteroids (Zentiva a.s.). The patient
was postoperatively treated for 10 days with Tamiflu (F.
Hoffmann-La Roche Ag) due to a positive test for influenza AB in a
donor. In the first EMB, there were histological findings without
evidence of cellular or antibody-mediated rejection.
Echocardiography showed good heart graft function and insignificant
pericardial effusion, without progression. Cardiopulmonary
compensation during hospitalization was without heart rhythm
disturbance. Donor 1 (male; age, 36 years) had suffered from
epilepsy since childhood and had suffered brain death due to
post-hypoxic brain damage after an epileptic seizure.
Patient 2 (female; age, 65 years) underwent OHT in
October 2017 at the Cardio Center of the Institute for Clinical and
Experimental Medicine (Prague, Czech Republic) due to congenital
cardiac malformation. The patient had been affected by an atrial
septal defect and underwent surgical closure in 1959 and
reoperation (surgical closure with bovine pericardial patch and
tricuspid valve plastic surgery) in 2011. In 2014, the patient had
been hospitalized multiple times due to right-sided heart failure.
In 2016, treatment with sildenafil (Mylan) was started because of
the detection of significant pulmonary arterial hypertension. In
March 2017, the patient was added to the waiting list for HTx. The
patient suffered from comorbidities such as permanent atrial
fibrillation, diabetes mellitus and dyslipidemia. Up to the 7th
postoperative day, the patient was supported with inotropic
milrinone for right-sided decompensation due to problematic
borderline diuresis with high furosemide (FSM; Zentiva a.s.)
support/preoperatively administered FSM at a dose of 1 g per day
for right-sided failure, without the need for an elimination
method. The patient received the same induction with rATG
(Fresenius) and maintenance therapy as patient 1. Corticoid-induced
deterioration of diabetes was compensated for by insulin therapy.
The first three EMBs showed histological findings without evidence
of cellular or antibody-mediated rejection. Echocardiography showed
a good heart graft with maximum moderate tissue relaxation with
inversion recovery and mild right ventricular dysfunction and
treated arterial hypertension. Donor 2 (female; age, 50 years) had
suffered from hypertension, hypertensive encephalopathy and
hypothyroidism, and brain death had occurred as a result of
intracerebral hemorrhage.
DNA analysis
Aortic tissues for genomic DNA extraction were
collected during OHT from donors and recipients from March 2016 to
October 2017 at the Institute for Clinical and Experimental
Medicine (Prague, Czech Republic). The screening of 48
single-nucleotide polymorphisms (SNPs; minor allele frequency range
0.4-0.5; Table SI) for
identification of the homozygous status was performed using
microfluidic chips (Fluidigm; cat. no. 48.48 IFC) on a
BioMark™ system (Fluidigm). The protocol was performed
according to manufacturer's recommendations stated in the Fluidigm
SNP Genotyping User Guide (PN68000098 REV.18, Appendix C-SNP
Type™ Assays for SNP Genotyping on the Dynamic
Array™ IFCs).
Blood samples (10 ml) were collected (before their
corresponding biopsy) in EDTA-containing tubes on the 10th day and
at the end of the 1st, 6th and 12th month after OHT (from April
2016 to October 2018 at the Cardio Center of the Institute for
Clinical and Experimental Medicine, Prague, Czech Republic), at the
times when EMB was performed. Plasma was separated (centrifugation
at 1,500 x g, 15 min, room temperature) within 30 min of blood
collection and stored at -80˚C in RNase-/DNase-free tubes. cfDNA
was extracted from 1 ml of plasma using a Plasma/Serum Cell-Free
Circulating DNA Purification MIDI kit (cat. no. 55600; Norgen
Biotek). DNA was eluted in a final volume of 45 µl. No artificial
spike was used.
The experiment was performed according the protocol
of Beck et al (16) with
slight modifications. Preamplification of the cfDNA was conducted
using the NEBNext Ultra II DNA Library Prep Kit (New England
Biolabs) with a final elution volume of 21 µl. Quantification of
cfDNA after this step was performed using a Qubit 4 fluorimeter
(Thermo Fisher Scientific, Inc.).
The total cfDNA concentration was determined using a
droplet PCR assay. PCR oligonucleotides and fluorophore-conjugated
hydrolysis probes were purchased from GeneriBiotech. ddPCR was
performed using a QX200 Droplet DPCR System (Bio-Rad Laboratories,
Inc.) in a volume of 20 µl containing 10 µl 2x ddPCR Supermix for
probes (no dUTP) (Bio-Rad Laboratories, Inc.), 7 µl of preamplified
DNA, 1.8 µl of primer mix (forward primer:
5'-TAGGCCATAATACTCTTGA-3'; reverse primer:
5'-ACTGGCATTCTAACTAGA-3'), 0.5 µl of FAM-labeled probe
(5'-TTGACATTTGGCCATTTTATAGGTCCA-3', and 0.5 µl of HEX-labeled probe
(5'-TGACATTTGGGCATTTTATAGGTCCA-3'), according to the manufacturer's
instructions. The thermocycling conditions were 95˚C for 10 min and
40 cycles of 94˚C for 30 sec and 57˚C for 1 min, followed by one
final step at 98˚C for 10 min. Sample analysis of each experiment
was performed using QuantaSoft v1.7 SW (Bio-Rad Laboratories,
Inc.). Rare event detection settings were used to calculate the
positive droplet concentration. The fluorescence threshold was set
manually based on validation data obtained from homozygous variants
of both versions, the temperature gradient and the negativity of
no-template controls. All reactions were performed in duplicate. To
account for the random distribution of target DNA into partitions,
Poisson's statistical model was applied and the absolute quantity
was calculated. Fractional abundance (%ddcfDNA) was calculated as
the ratio of donor absolute quantity [copies (cp) per 1 ml] to
absolute quantity of cfDNA (donor absolute quantity plus recipient
absolute quantity).
Conversion of ddPCR results
The ddPCR concentration results were converted to a
plasma-equivalent concentration (cp/ml) using the following
equation:
where Cplasma is the concentration
of the target within the plasma in cp/ml; CddPCR
is the concentration of the target within the PCR in cp/µl;
Velution is the volume of eluent used during the
cfDNA extraction step in µl; VcfDNA is the volume
of extracted cfDNA used in the PCR in µl; VMM is
the volume of all other PCR components in µl; and
Vplasma is the volume of plasma used for cfDNA
extraction in ml.
Demonstration of ddcfDNA measurement
on two digital PCR instruments
For comparing the quantification of ddcfDNA, two
digital PCR systems were used, namely the QX200 digital droplet PCR
system (Bio-Rad Laboratories) and the Qiacuity One dPCR system
(5plex; Qiagen GmbH). The measurements were conducted on samples of
patient 2. The first method used for Bio-Rad is provided in the
Methods section. For the second measurement, Qiacuity was used,
which involved a volume of 40 µl with 10 µl of 4X Probe PCR Master
Mix (Qiagen GmbH), 8.8 µl of preamplified DNA, 4 µl of 10X
primer-probe mix (0.8 µM forward primer: 5'-TAGGCCATAATACTCTTGA-3';
0.8 µM reverse primer: 5'-ACTGGCATTCTAACTAGA-3'; 0.4 µM FAM-labeled
probe (5'-TTGACATTTGGCCATTTTATAGGTCCA-3') and 4 µl of 10X
primer-probe mix (0.8 µM forward primer: 5'-TAGGCCATAATACTCTTGA-3';
0.8 µM reverse primer: 5'-ACTGGCATTCTAACTAGA-3'; 0.4 µM HEX-labeled
probe (5'-TGACATTTGGGCATTTTATAGGTCCA-3') and 13.2 µl of RNAse free
water, following the manufacturer's instructions. The thermocycling
conditions were 95˚C for 2 min and 40 cycles of 95˚C for 15 sec and
60˚C for 30 min. Quiacuity Nanoplates 26k (Qiagen GmbH) with up to
26,000 partitions per well were used to conduct the measurements.
The ddcfDNA absolute quantity and %ddcfDNA were calculated, and the
Bland-Altman analysis was performed using GraphPad Prism 5
software, version 5.03 (GraphPad; Dotmatics) (17).
Results
SNP screening
SNP screening identified 10 (patient 1) and 7
(patient 2) different homozygous variants suitable for the
detection of donor DNA in paired samples (Table SI). All selected SNP primers (for
sequences see Table SI) were
validated before measurement to eliminate false-positive droplets.
Only variant rs521861 (within the MYO5B gene) passed
validation for using the ddPCR method. Target sequences C/G within
rs521861 were analyzed in plasma cfDNA of the patients.
Case reports
In patient 1, OHT was performed without any
complications and the postoperative course was favorable.
Echocardiographic parameters were normal except for mild
dysfunction of the dilated right ventricle during the first days
after Tx. Increased wall thickness was detected and was treated
with intravenous (IV) pulse dose steroids. Other events were not
observed during follow-up. EMB examination confirmed mild grades of
ACR (1R/2) in the 1st month and (1R/1A) in the 6th month after OHT.
The maximal ddcfDNA level of 270 cp/ml was detected in the 1st
month after Tx. A decline in ddcfDNA to 80 cp/ml was then detected
in the 6th month after OHT and 53 cp/ml (time without rejection;
Fig. 1A). At the time of conclusion
of the present study (October 2023), patient 1 is free of any
cardiac problems and does not have any coronary graft disease.
 | Figure 1Quantity of ddcfDNA (copies/ml). (A)
Patient 1 with confirmed ACR and (B) patient 2 with confirmed AMR.
0R, ACR and AMR grade 0; 10D, 10th day; 1M, at the end of the 1st
month; 6M, at the end of the 6th month; 12M, at the end of the 12th
month after orthotopic heart Tx; AMR, antibody-mediated rejection;
ACR, acute cellular rejection; ddcfDNA, donor-derived cell-free
DNA; cp, copies; Tx, transplantation. |
In patient 2, the OHT was performed without any
complications. Right-sided heart decompensation was treated with
inotropic drugs (milrinone) until the 6th postoperative day.
Echocardiography showed dilatation and dysfunction of the right
ventricle and secondary tricuspid regurgitation, while all other
parameters were normal. At one month after HTx, the right ventricle
had a standard size and only mild dysfunction, but the wall
thickness was detected to increase. EMBs were without any
histological signs of acute cellular or humoral rejection at this
point. In the 2nd month after transplantation, an oral pulse dose
of steroids was indicated due to mild acute cellular rejection
(grade 1R/2). Echocardiographic parameters were normal. EMB
examination confirmed moderate-grade ACR (2R/3A) in the 3rd month
after OHT, which was treated with 1,000 mg/day of IV
methylprednisolone for 3 days. EMB examination at the end of the
6th month after OHT confirmed AMR grade 1+. Due to the histological
findings of AMR, complement-dependent cytotoxicity (CDC) crossmatch
and human leukocyte antigen antibody (HLA; Luminex® bead
assay; Luminex Corp.) testing were added; this was performed
according to standard procedures. The specificity of HLA antibodies
was defined by LABScreen Mixed and Single Antigen class I and class
II beads (One Lambda Inc.). CDC crossmatch was negative. Only
non-significant positivity of anti-HLA class II antibodies (anti-DR
13, anti-DR 14 and anti-DR 17; One Lambda Inc.) was detected. In
view of the above, the tacrolimus dose was increased and a higher
target tacrolimus level was set. Echocardiography showed only a
mild thickening of the interventricular septum (14 mm) without
deterioration of ventricular function. The patient was treated with
Metroprolol succinate (Astra Zeneca Spa), Perindoprilum
Argininum/Amlodipinum (Servier Industries Ltd.) and FSM. This was
the last time when a rejection episode was detected. Corresponding
to the EMB results, a slight increase of ddcfDNA 1,416 cp/ml was
identified early in the sample collected 1 month after OHT, without
serious rejection. The highest value of ddcfDNA at 1,846 cp/ml was
detected 6 months after transplantation when AMR grade 1+ was also
detected. Subsequently, there was a decline of ddcfDNA to 777 cp/ml
in the rejection-free time (Fig.
1B). At the time of conclusion of the present study (October
2023), patient 2 is in a stable clinical condition, but is
classified as New York Heart Association functional class III.
After transplantation, each patient is monitored and
treated until death in our institute. Regular medical follow-ups
are scheduled every 6 months to assess the status and function of
the graft.
Comparative analysis
The ddcfDNA measurements obtained using two
different digital PCR instruments were compared. There was a
similar trend in the %ddcfDNA and in the plasma quantity of ddcfDNA
(Table II and Fig. 2). The slight differences in results
may have been influenced by the different PCR reagents, efficiency
and specificity of the instruments.
 | Table IIComparison of measurements using two
different digital PCR instruments. |
Table II
Comparison of measurements using two
different digital PCR instruments.
| A, ddcfDNA, % |
|---|
| Time post-OHT | Bio-Rad | Qiacuity | Average | Difference | Bias ± SD | 95% Limit of
agreement |
|---|
| 10D | 0.6772 | 0.731 | 0.704 | -0.054 | -1.866±2.288 | -6.350 to 2.618 |
| 1M | 2.6074 | 4.646 | 3.627 | -2.038 | | |
| 6M | 5.1292 | 10.164 | 7.647 | -5.035 | | |
| 12M | 0.5829 | 0.918 | 0.751 | -0.335 | | |
| B, ddcfDNA,
cp/ml |
| Time post-OHT | Bio-Rad | Qiacuity | Average | Difference | Bias ± SD | 95% Limit of
agreement |
| 10D | 291.6 | 147.54 | 219.57 | 144.06 | -377.1±578.6 | -1511 to 756.9 |
| 1M | 1416.34 | 2612.25 | 2014.30 | -1195.91 | | |
| 6M | 1846.8 | 2169.64 | 2008.22 | -322.84 | | |
| 12M | 777.8 | 911.25 | 844.43 | -133.65 | | |
Discussion
Gentle detection of acute rejection is one of the
central tasks in transplantology. The standard invasive method is
EMB, which is carried out using a vessel to catheterize the heart,
which takes 3-4 biopsies from the right ventricle, and then graded
histologically. All patients undergo at least 8-10 biopsies in the
first year after transplantation, most within the first 3 months.
The presence of cell-free human DNA in different body fluids gives
new opportunities in the diagnosis of (not only) graft rejection.
The release of ddcfDNA into recipients' blood due to myocardial
cell damage makes these molecules potential biomarkers of graft
health. The ddcfDNA kinetics seem to follow an L-shaped curve with
high concentration immediately after transplantation, decreasing
over a week to a stable level (6).
CfDNA level monitoring using the newly established and more
sensitive methods (such as ddPCR) may contribute to the early
detection of allograft rejection.
In the present case study, the value of the ddPCR
method combined with selected SNP typing for monitoring rejection
was verified. The dynamics of ddcfDNA between the 1st and 6th
months post-Tx reflected cardiac graft injury in both patients. A
ddcfDNA fractional abundance of 0.84% was found during confirmed
ACR (Fig. 3A) and 5.1% during
confirmed AMR (Fig. 3B), which may
well be in agreement with previous findings (18).
 | Figure 3ddcfDNA fractional determination (%).
(A) Patient 1 with confirmed ACR and (B) patient 2 with confirmed
AMR. The fractional abundance (%ddcfDNA) was calculated as the
ratio of donor absolute quantity (cp/ml) to absolute quantity of
cfDNA (donor absolute quantity plus recipient absolute quantity).
0R, ACR and AMR grade 0; 10D, 10th day; 1M, at the end of the 1st
month; 6M, at the end of the 6th month; 12M, at the end of the 12th
month after orthotopic heart Tx; ddcfDNA, donor-derived cell-free
DNA; cp, copies; Tx, transplantation; AMR, antibody-mediated
rejection; ACR, acute cellular rejection. |
In patient 2, EMB examination confirmed mild- and
moderate-grade ACR (1R/2 and 2R/3A) and mild right ventricular
dysfunction within the time between the 1st and 6th months after
Tx. Higher levels of ddcfDNA were able to reflect graft injury, as
at the same time-points, increased levels of B-type natriuretic
peptide (BNP); 1M=894 ng/l; 6M=402 ng/l and 12M=275 ng/l were
detected. BNP is a neurohormone secreted from cardiac ventricles in
response to ventricular strain (19). The BNP titer may be influenced by
severe rejection episodes and diastolic dysfunction, and possibly
intracardiac pressure derangement (20). Plasma BNP >90 pg/ml may serve as
a marker for ventricular dysfunction. Elevated BNP was also
associated with Grade ≥2 rejection (19,20).
Previous studies have described the posttransplant
decay kinetics of ddcfDNA. Elevated median levels of ddcfDNA
(approximately 4% in heart, 10% in renal, 26% in lung and 70% in
liver Tx) within the first few days after surgery most likely
reflect ischemia/reperfusion damage in the graft related to the
transplant process. Within the first 10 days after Tx, in stable
patients with no graft injury, the mean %ddcfDNA may decrease to a
level below 0.06-10%, depending on the type of signs of
transplanted organ (4,9,18,21,22).
Agbor-Enoh et al (18)
detected an increase in %ddcfDNA at 0.5 and 3.2 months before the
histopathological diagnosis of ACR or AMR. Furthermore, in this
study (18), the %ddcfDNA showed
distinctive characteristics that varied between AMR and ACR.
In the present study, only two cases with different
acute rejection were analyzed. The primary focus was on the trend
of ddcfDNA in comparison to EMB results within the post-Tx
time-points. However, the absence of statistical analyses may be a
limitation of the current study. To identify the optimal threshold,
a larger cohort needs to be measured. The limitation of this method
may be the small amount of ddcfDNA in plasma. In addition,
increased release of recipient DNA into the bloodstream due to
infections, exercise, medications or non-graft-associated vascular
compromise may influence the lowering of %ddcfDNA (9). Using a preamplification step and
absolute quantification (as copies per ml plasma), which are not
affected by changes in recipient cfDNA, may avoid such biases.
The comparison of measurements obtained from two
different digital PCRs is limited in the present study due to the
lack of results from patient 1. To confirm the present findings, it
is necessary to measure more genetic variants in a larger number of
patients.
In the present case study, ddcfDNA was successfully
measured as a marker for acute rejection in two patients. However,
the present results need to undergo verification in a larger cohort
to validate the reliability and generalizability of ddcfDNA as a
biomarker.
In conclusion, in the present study, increased
levels of ddcfDNA were detected during ongoing ACR and AMR.
Individual monitoring of ddcfDNA dynamics from the 1st to the 6th-
month post-Tx reflected cardiac graft injury in patients suffering
ACR or AMR, meaning ddcfDNA may serve as a noninvasive
biomarker.
Supplementary Material
List of 48 single-nucleotide
polymorphisms selected for screening.
Acknowledgements
The article is based on an abstract presented as a
poster at the XXXI Congress of the Czech Society of Cardiology from
May 14 to 16, 2023, in Brno (Czech Republic) and published online
as abstract no. 359(23).
Funding
Funding: This work was supported the Ministry of Health of the
Czech Republic- DRO ('Institute of Clinical and Experimental
Medicine - IKEM, IN 00023001') and the Ministry of Health of the
Czech Republic (grant no. NU20-06-00061).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
The original draft was written by DD and JAH. JV and
DD designed and directed the study, summarized the clinical
characteristics, contributed to the interpretation of the results
and edited the article. ER and PH designed and performed the
experiments, analyzed the data and wrote the article. DD and ER saw
and verified all raw data. SN coordinated and created clinical
databases. Data review was performed by JAH. All authors reviewed
the original draft and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study was approved on 15 June 2015 (approval no.
G-15-06-15) and on 12 June 2019 (approval no. G-19-37), both
granted by the Ethics Committee of the Institute for Clinical and
Experimental Medicine and Thomayer Hospital with Multicenter
Competence in Prague, Czech Republic. Two ethics approvals are
included because patients were included in two different studies.
The second study, which focuses on circulating microRNA as
biomarkers of graft dysfunction, has not yet been published.
Patient consent for publication
The study was performed according to the Declaration
of Helsinki (2000) of the World Medical Association (24). Both patients provided written
informed consent regarding analyses and examinations, as well as
publication of case descriptions and results.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Otto MEB, Martins AMA, Campos Dall'Orto
AOM, Leite SF, de Queiroz Mauricio Filho MAF, Martins NT, de Araújo
SR, Almeida SV, Paiva MUB and Atik FA: Acute cellular rejection in
heart transplant patients: Insights of global longitudinal strain,
myocardial work, and an exclusive group of chagas disease. Front
Cardiovasc Med. 9(841698)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Suzuki N, Kamataki A, Yamaki J and Homma
Y: Characterization of circulating DNA in healthy human plasma.
Clin Chim Acta. 387:55–58. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Beck J, Bierau S, Balzer S, Andag R,
Kanzow P, Schmitz J, Gaedcke J, Moerer O, Slotta JE, Walson P, et
al: Digital droplet PCR for rapid quantification of donor DNA in
the circulation of transplant recipients as a potential universal
biomarker of graft injury. Clin Chem. 59:1732–1741. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schütz E, Fischer A, Beck J, Harden M,
Koch M, Wuensch T, Stockmann M, Nashan B, Kollmar O, Matthaei J, et
al: Graft-derived cell-free DNA, a noninvasive early rejection and
graft damage marker in liver transplantation: A prospective,
observational, multicenter cohort study. PLoS Med.
14(e1002286)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kustanovich A, Schwartz R, Peretz T and
Grinshpun A: Life and death of circulating cell-free DNA. Cancer
Biol Ther. 20:1057–1067. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
De Vlaminck I, Valantine HA, Snyder TM,
Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Bernstein D,
Weisshaar D, et al: Circulating cell-free DNA enables noninvasive
diagnosis of heart transplant rejection. Sci Transl Med.
6(241ra77)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hidestrand M, Tomita-Mitchell A,
Hidestrand PM, Oliphant A, Goetsch M, Stamm K, Liang HL,
Castleberry C, Benson DW, Stendahl G, et al: Highly sensitive
noninvasive cardiac transplant rejection monitoring using targeted
quantification of donor-specific cell-free deoxyribonucleic acid. J
Am Coll Cardiol. 63:1224–1226. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Whitlam JB, Ling L, Skene A, Kanellis J,
Ierino FL, Slater HR, Bruno DL and Power DA: Diagnostic application
of kidney allograft-derived absolute cell-free DNA levels during
transplant dysfunction. Am J Transplant. 19:1037–1049.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Oellerich M, Shipkova M, Asendorf T,
Walson PD, Schauerte V, Mettenmeyer N, Kabakchiev M, Hasche G,
Gröne HJ, Friede T, et al: Absolute quantification of donor-derived
cell-free DNA as a marker of rejection and graft injury in kidney
transplantation: Results from a prospective observational study. Am
J Transplant. 19:3087–3099. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Oellerich M, Budde K, Osmanodja B,
Bornemann-Kolatzki K, Beck J, Schütz E and Walson PD: Donor-derived
cell-free DNA as a diagnostic tool in transplantation. Front Genet.
13(1031894)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Beck J, Oellerich M, Schulz U, Schauerte
V, Reinhard L, Fuchs U, Knabbe C, Zittermann A, Olbricht C, Gummert
JF, et al: Donor-derived cell-free DNA is a novel universal
biomarker for allograft rejection in solid organ transplantation.
Transplant Proc. 47:2400–2403. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gielis EM, Beirnaert C, Dendooven A,
Meysman P, Laukens K, De Schrijver J, Van Laecke S, Van Biesen W,
Emonds MP, De Winter BY, et al: Plasma donor-derived cell-free DNA
kinetics after kidney transplantation using a single tube multiplex
PCR assay. PLoS One. 13(e0208207)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Berry GJ, Burke MM, Andersen C, Bruneval
P, Fedrigo M, Fishbein MC, Goddard M, Hammond EH, Leone O, Marboe
C, et al: The 2013 international society for heart and lung
transplantation working formulation for the standardization of
nomenclature in the pathologic diagnosis of antibody-mediated
rejection in heart transplantation. J Heart Lung Transplant.
32:1147–1162. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Stewart S, Winters GL, Fishbein MC,
Tazelaar HD, Kobashigawa J, Abrams J, Andersen CB, Angelini A,
Berry GJ, Burke MM, et al: Revision of the 1990 working formulation
for the standardization of nomenclature in the diagnosis of heart
rejection. J Heart Lung Transplant. 24:1710–1720. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cooper LT, Baughman KL, Feldman AM,
Frustaci A, Jessup M, Kuhl U, Levine GN, Narula J, Starling RC,
Towbin J, et al: The role of endomyocardial biopsy in the
management of cardiovascular disease: A scientific statement from
the American heart association, the American college of cardiology,
and the European society of cardiology. Circulation. 116:2216–2233.
2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Beck J, Oellerich M and Schütz E: A
universal droplet digital PCR approach for monitoring of graft
health after transplantation using a preselected SNP set. Methods
Mol Biol. 1768:335–348. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Altman DG and Bland JM: Measurement in
medicine: The analysis of method comparison studies. Statistician.
32:307–317. 1983.
|
|
18
|
Agbor-Enoh S, Shah P, Tunc I, Hsu S,
Russell S, Feller E, Shah K, Rodrigo ME, Najjar SS, Kong H, et al:
Cell-free DNA to detect heart allograft acute rejection.
Circulation. 143:1184–1197. 2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yardan T, Altintop L, Baydin A, Yilmaz O
and Guven H: B-type natriuretic peptide as an indicator of right
ventricular dysfunction in acute pulmonary embolism. Int J Clin
Pract. 62:1177–1182. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lan YT, Chang RKR, Alejos JC, Burch C and
Wetzel GT: B-type natriuretic peptide in children after cardiac
transplantation. J Heart Lung Transplant. 23:558–563.
2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gielis EM, Ledeganck KJ, Dendooven A,
Meysman P, Beirnaert C, Laukens K, De Schrijver J, Van Laecke S,
Van Biesen W, Emonds MP, et al: The use of plasma donor-derived,
cell-free DNA to monitor acute rejection after kidney
transplantation. Nephrol Dial Transplant. 35:714–721.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Macher HC, García-Fernández N,
Adsuar-Gómez A, Porras-López M, González-Calle A, Noval-Padillo J,
Guerrero JM, Molinero P, Borrego-Domínguez JM, Herruzo-Avilés Á and
Rubio A: Donor-specific circulating cell free DNA as a noninvasive
biomarker of graft injury in heart transplantation. Clin Chim Acta.
495:590–597. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
(https://www.cksonline.cz/31-vyrocni-sjezd-cks/sjezd.php?p=read_abstrakt_program&idabstrakta=370&act=print#:~:text=Conclusion%3A%20Individual%20monitoring%20of%20ddcfDNA,patients%20suffering%20ACR%20or%20AMR.
|
|
24
|
World Medical Association. World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|