Introduction
The innate immune system is recognized as the body's
first line of defense against pathogens, engaging in the immediate
destruction and elimination of pathogens during the initial stages
of infection (1). This primary
response mechanism is crucial for preventing the establishment and
spread of infectious agents (1).
Macrophages are recognized as pivotal phagocytes within the innate
immune system, playing a crucial role in mediating the initial
immune response to pathogens (2).
The primary function of macrophages is the phagocytosis of foreign
pathogens that invade the human body (3). It is reported that through the process
of engulfing these foreign pathogens, macrophages generate a
variety of immunostimulatory factors such as nitric oxide (NO),
prostaglandin E2 (PGE2), inducible nitric oxide synthase (iNOS),
cyclooxygenase-2 (COX-2), IL-1β and TNF-α (4). It is well-documented that various
immunostimulatory factors secreted by activated macrophages play a
crucial role in the activation of adaptive immune cells, including
T cells and B cells (2).
Furthermore, macrophages are known to possess a pivotal
antigen-presenting capability that is essential for the initiation
of the adaptive immune response against invading pathogens
(5). Consequently, macrophages are
reported to be crucial in both innate and adaptive immune responses
(6). Thus, research is currently
underway to screen various natural agents capable of activating
macrophages, thereby simultaneously enhancing both innate and
adaptive immune responses within the body (7-9).
Compared to other species of Sambucus,
Sambucus racemosa subsp. pendula (SRP) is
distinguished by its smaller berries and unique inflorescence
morphology (10). This species is
endemic to Korea and is known to be exclusively distributed on
Ulleungdo Island (10). The berries
of Sambucus have been widely used not only in foods such as
jams and juices but also for medicinal purposes (10). However, the Korean Ministry of Food
and Drug Safety permits only the leaves and shoots of SRP to be
used as food. Nevertheless, studies on the pharmacological effects
of leaves and shoots of SRP remain nonexistent. Elderberry,
specifically from the Sambucus genus (Sambucus
nigra), has been studied for its immune-boosting properties
(11). Thus, the present study
investigated the immunostimulatory activity of extracts from the
leaves of SRP in RAW264.7 macrophage cells to investigate their
potential use as immuno-enhancing products.
Materials and methods
Chemical reagents
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT; cat. no. 475989), PD98059 (cat. no. 513000), SB203580
(cat. no. 58307), SP600125 (cat. no. 55567), TAK-242 (cat. no.
614316), Neutral Red (cat. no. N4638) and Griess reagent (cat. no.
G4410) were purchased from MilliporeSigma. C29 (cat. no. 27029) was
purchased from Cayman Chemical Company. The primary antibodies such
as phosphorylated (p-)JNK (cat. no. 9251), JNK (cat. no. 9258) and
β-actin (cat. no. 5125) and the secondary antibody for anti-rabbit
IgG, HRP-linked antibody (cat. no. 7074) was purchased from Cell
Signaling Technology, Inc.
Sample preparation
Leaves of Sambucus racemosa subsp.
pendula (SRPL; voucher number: FMRC-230501A1-A3) were
collected from Ulleungdo Island, Korea in May 2023. SPRL was
taxonomically identified by the Forest Medicinal Resources Research
Center (Yeongju, South Korea) before being provided for the present
study. Ginseng was also provided from the Forest Medicinal
Resources Research Center. The freeze-dried SRPL was subjected to
an immersion extraction at 20˚C for 24 h using 100% distilled
water, 30% ethanol, 50% ethanol and 70% ethanol in a 20-fold volume
ratio. Additionally, the freeze-dried SPRL were subjected to
immersion extraction with 100% distilled water in a 20-fold volume
ratio at 20, 40, 60 and 80˚C for 24 h. The freeze-dried Ginseng
(AE20-G) was subjected to immersion extraction with 100% distilled
water in a 20-fold volume ratio at 20˚C for 24 h. After 24 h, each
extract was centrifuged at 25,200 x g for 10 min at 4˚C and then
freeze-dried. The freeze-dried extracts obtained with 100%
distilled water were re-dissolved in distilled water, while the
freeze-dried ethanol extracts were re-dissolved in dimethyl
sulfoxide (DMSO) for use in the experiments.
Cell culture
RAW264.7 murine macrophages (cat. no. TIB-71) were
sourced from the American Type Culture Collection. RAW264.7 cells
were cultivated in a CO2 incubator maintained at 37˚C
with 5% CO2 using Dulbecco's Modified Eagle Medium/F-12
(DMEM/F-12; cat. no. SH3023.01; Cytiva) supplemented with 10% fetal
bovine serum (FBS; cat. no. 16000-044; Gibco; Thermo Fisher
Scientific, Inc.), penicillin (100 units/ml) and streptomycin (100
µg/ml).
Measurement of the viability of
RAW264.7 cells
The cytotoxicity of the samples on RAW264.7 cells
was evaluated using the MTT assay. RAW264.7 cells (1x105
cells/well) were cultured in a 96-well plate at 37˚C for 24 h,
followed by treatment with the samples (6.25-200 µg/ml) at various
concentrations and further incubation at 37˚C for 24 h. The control
group (CON) did not undergo any treatment. Subsequently, MTT
solution (1 mg/ml) was added to each well and the cells were
incubated at 37˚C for an additional 4 h. After this incubation
period, the culture medium was removed and the crystallized MTT was
dissolved in DMSO. The absorbance was then measured at 570 nm using
a UV/visible spectrophotometer (Xma-3000PC; Human Corporation).
Measurement of NO and PGE2 levels in
RAW264.7 cells
RAW264.7 cells (1x105 cells/well) were
cultured in a 96-well plate at 37˚C for 24 h, followed by treatment
with the samples (6.25-200 µg/ml) at various concentrations and
incubation at 37˚C for another 24 h. In addition, RAW264.7 cells
were pre-treated with C29 (100 µM), TAK-242 (10 µM), PD98059 (20
µM), SB203580 (20 µM) or SP600125 (20 µM) at 37˚C for 2 h and then
co-treated with the sample (50 µg/ml) at 37˚C for 24 h. RAW264.7
cells were cultured in a 96-well plate at 37˚C for 24 h, followed
by treatment with AE20-SRPL (50 µg/ml) or AE20-G (50 µg/ml) and
incubation at 37˚C for another 24 h. After this incubation period,
NO levels were measured using the Griess assay. The cell culture
supernatant was mixed with Griess reagent in a 1:1 ratio and
allowed to react at room temperature for 15 min. The absorbance was
then measured at 540 nm using a UV/Visible spectrophotometer
(Xma-3000PC; Human Corporation). PGE2 levels were determined using
a Mouse Prostaglandin E2 (PGE2) ELISA Kit (cat. no. MBS266212;
MyBioSource, Inc.) according to the protocols provided by the
manufacturer.
Measurement of phagocytotic activity
in RAW264.7 cells
The effect of the samples on phagocytosis in
RAW264.7 cells was evaluated using the Neutral Red uptake assay.
RAW264.7 cells (1x105 cells/well) were cultured in a
96-well plate at 37˚C for 24 h, followed by treatment with the
samples (12.5-50 µg/ml) and further incubation at 37˚C for 24 h. In
addition, RAW264.7 cells were pre-treated with TAK-242 (10 µM) or
SP600125 (20 µM) at 37˚C for 2 h and then co-treated with the
sample (50 µg/ml) at 37˚C for 24 h. After this period, the cells
were stained with 0.01% Neutral Red solution at 37˚C for 2 h. The
stained Neutral Red was then extracted from the cells using a lysis
buffer (50% ethanol/1% acetic acid). The absorbance was
subsequently measured at 540 nm using a UV/Visible
spectrophotometer (Xma-3000PC; Human Corporation).
Reverse transcription-quantitative
(RT-q)PCR
RAW264.7 cells (2x106 cells/well) were
cultured in a 6-well plate at 37˚C for 24 h, followed by treatment
with the samples (12.5-50 µg/ml) and further incubation at 37˚C for
24 h. In addition, RAW264.7 cells were pre-treated with C29 (100
µM), TAK-242 (10 µM), PD98059 (20 µM), SB203580 (20 µM) or SP600125
(20 µM) at 37˚C for 2 h and then co-treated with the sample (50
µg/ml) at 37˚C for 24 h. Following all treatments, total RNA was
isolated from RAW264.7 cells using the RNeasy Mini Kit (cat. no.
74104; Qiagen GmbH) according to the manufacturer's protocol.
Subsequently, cDNA was synthesized from 1 µg of total RNA using the
Verso cDNA Kit (cat. no. AB1453A; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RT-qPCR was performed
using a Rotor-Gene Q (cat. no. 9001862; Qiagen GmbH) with a
QuantiTect SYBR Green PCR kit (cat. no. 204143; Qiagen GmbH) and
primers according to the manufacturer's protocol. RT-qPCR cycling
conditions were as follows: initial denaturation at 95˚C for 5 min,
30 cycles of denaturation at 95˚C for 15 sec, annealing at 55˚C for
30 sec, and extension at 72˚C for 30 sec to 1 min, followed by a
final extension at 72˚C for 10 min, with an optional hold at 4˚C.
Data analysis was conducted using Rotor-Gene Q Series software
2.3.5 (Qiagen GmbH). Transcription levels were normalized to those
of the GAPDH gene. The formula used to analyze mRNA expression was
2-IICq, where
IICq=(Cttarget-CtGAPDH)sample-(Cttarget-CtGAPDH)control
(12). The sequences of the primers
used in the present study were as follows: iNOS mRNA, Forward:
5'-GTTACCATGAGGCTGAAATCC-3' and Reverse:
5'-CCTCTTGTCTTTGACCCAGTAC-3'; COX-2 mRNA, Forward:
5'-CTGGAACATGGACTCACTCAGTTTG-3' and Reverse:
5'-AGGCCTTTGCCACTGCTTGT-3'; IL-1β mRNA, Forward:
5'-GGTACATCAGCACCTCAC-3' and Reverse: 5'-AAACAGTCCAGCCCATAC-3';
TNF-α mRNA, F: 5'-CTCTTCTCATTCCTGCTTG-3' and Reverse:
5'-CTCCACTTGGTGGTTTGT-3'; GAPDH mRNA, Forward:
5'-GGACCTCATGGCCTACATGG-3' and Reverse:
5'-TAGGGCCTCTCTTGCTCAGT-3'.
SDS-PAGE and western blot
analysis
RAW264.7 cells (2x106 cells/well) were
cultured in a 6-well plate at 37˚C for 24 h. Then, RAW264.7 cells
were treated with the samples (50 µg/ml) at 37˚C for 1, 3, 6, 10,
or 24 h. In addition, RAW264.7 cells were pretreated with TAK-242
(10 µg/ml) at 37˚C for 2 h and then co-treated with the sample (50
µg/ml) at 37˚C for 1 h. After washing with phosphate-buffered
saline, proteins were extracted from RAW264.7 cells using a
radioimmunoprecipitation assay buffer (cat. no. BP-115DG; Boston
BioProducts, Inc.). The resulting lysates were centrifuged at 4˚C
and 25,200 x g for 30 min. Protein concentrations were quantified
using Pierce BCA Protein Assay Kits (cat. no. 23225; Thermo Fisher
Scientific, Inc.). The proteins (50 µg/well) were then separated
via SDS-PAGE (12% polyacrylamide gel) and subsequently transferred
to nitrocellulose membranes (0.45 µm; cat. no. 88018; Thermo Fisher
Scientific, Inc.). These membranes underwent a blocking step using
5% nonfat milk for 1 h at ambient temperature, followed by
overnight incubation with primary antibodies (dilution 1:1,000) at
4˚C. This was succeeded by a one-hour incubation with secondary
antibodies (dilution 1:1,000) at room temperature. After treatment
with ECL Select Western Blotting Detection Reagent (cat. no.
RPN2232; Cytiva), visualization of the protein bands was
accomplished using an LI-COR C-DiGit Blot Scanner (LI-COR
Biosciences). The intensity of these protein bands was
quantitatively determined using the UN-SCAN-IT gel software version
5.1 (Silk Scientific Inc.).
Statistical analysis
All experiments were repeated at least thrice.
Statistical analyses were conducted using GraphPad Prism version
5.0 (Dotmatics) and data are represented as the mean ± standard
deviation. All data were analyzed using one-way analysis of
variance, followed by Bonferroni's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
AE20-SRPL induces macrophage
activation in RAW264.7 cells
To investigate the role of SRPL in inducing
macrophage activation, we examined the changes in immunostimulatory
factors and phagocytic activity in RAW264.7 cells treated with
SRPL. To determine the optimal extraction condition for SRPL, a
comparative analysis of SRPL extracted by 100% distilled water
(AE-SRPL), 30% ethanol (30EE-SRPL), 50% ethanol (50EE-SRPL), or 70%
ethanol (70EE-SRPL) on NO production in RAW264.7 cells was
conducted. As shown in Fig. 1A,
AE-SRPL significantly induced NO production compared with the
control, demonstrating its potential as an immunostimulant, whereas
30EE-SRPL induced only a modest increase and 50EE-SRPL and
70EE-SRPL barely induced NO production. Therefore, to determine
whether the extraction temperature of SRPL affected NO production
in RAW264.7 cells, NO production in RAW264.7 cells treated with
SRPL extracts obtained at 20˚C (AE20-SRPL), 40˚C (AE40-SRPL), 60˚C
(AE60-SRPL) and 80˚C (AE80-SRPL) were compared and analyzed. As
shown in Fig. 1B, RAW264.7 cells
treated with AE20-SRPL and AE40-SRPL exhibited significant
increases in NO production. By contrast, AE60-SRPL and AE80-SRPL
did not induce any notable enhancement in NO production in these
cells. Given that AE20-SRPL induced a higher production of NO than
AE40-SRPL, AE20-SRPL was selected for further investigation. As
shown in Fig. 2A, AE20-SRPL
(6.25-200 µg/ml) elicited a statistically significant,
concentration-dependent increase in the production of NO and PGE2
in RAW264.7 cells. To determine whether the AE20-SRPL-mediated
increases in NO and PGE2 production were due to elevated expression
of iNOS and COX-2, respectively, the mRNA expression changes of
iNOS and COX-2 in RAW264.7 cells treated with AE20-SRPL were
analyzed using RT-qPCR analysis. As shown in Fig. 2B, AE20-SRPL (12.5-50 µg/ml)
significantly enhanced the mRNA expression of iNOS and COX-2.
Additionally, AE20-SRPL (12.5 µg/ml ~50 µg/ml) also elevated the
mRNA expression of IL-1β and TNF-α in RAW264.7 cells (Fig. 2C). To assess whether AE20-SRPL
activates phagocytic functions in macrophages, changes in
phagocytic activity in RAW264.7 cells treated with AE20-SRPL were
analyzed using the neutral red uptake assay. As shown in Fig. 2D, AE20-SRPL (12.5-50 µg/ml) was
found to activate phagocytic functions in RAW264.7 cells in a
concentration-dependent manner. Finally, the effect of AE20-SRPL on
cell viability in RAW264.7 cells was assessed using MTT assay. The
results indicated that AE20-SRPL, across a concentration range from
6.25-200 µg/ml, did not adversely affect the viability of RAW264.7
cells, indicating that AE20-SRPL is non-toxic to macrophages at the
tested concentrations (Fig. 2E).
Additionally, to ascertain the extent of macrophage activation
induced by AE20-SRPL, the NO production in RAW264.7 cells
stimulated by AE20-SRPL or the well-known immunostimulant ginseng
(AE20-G) were compared. As shown in Fig. 2F, in RAW264.7 cells stimulated with
AE20-SRPL, a greater increase in NO production was observed
compared with cells stimulated with AE20-G.
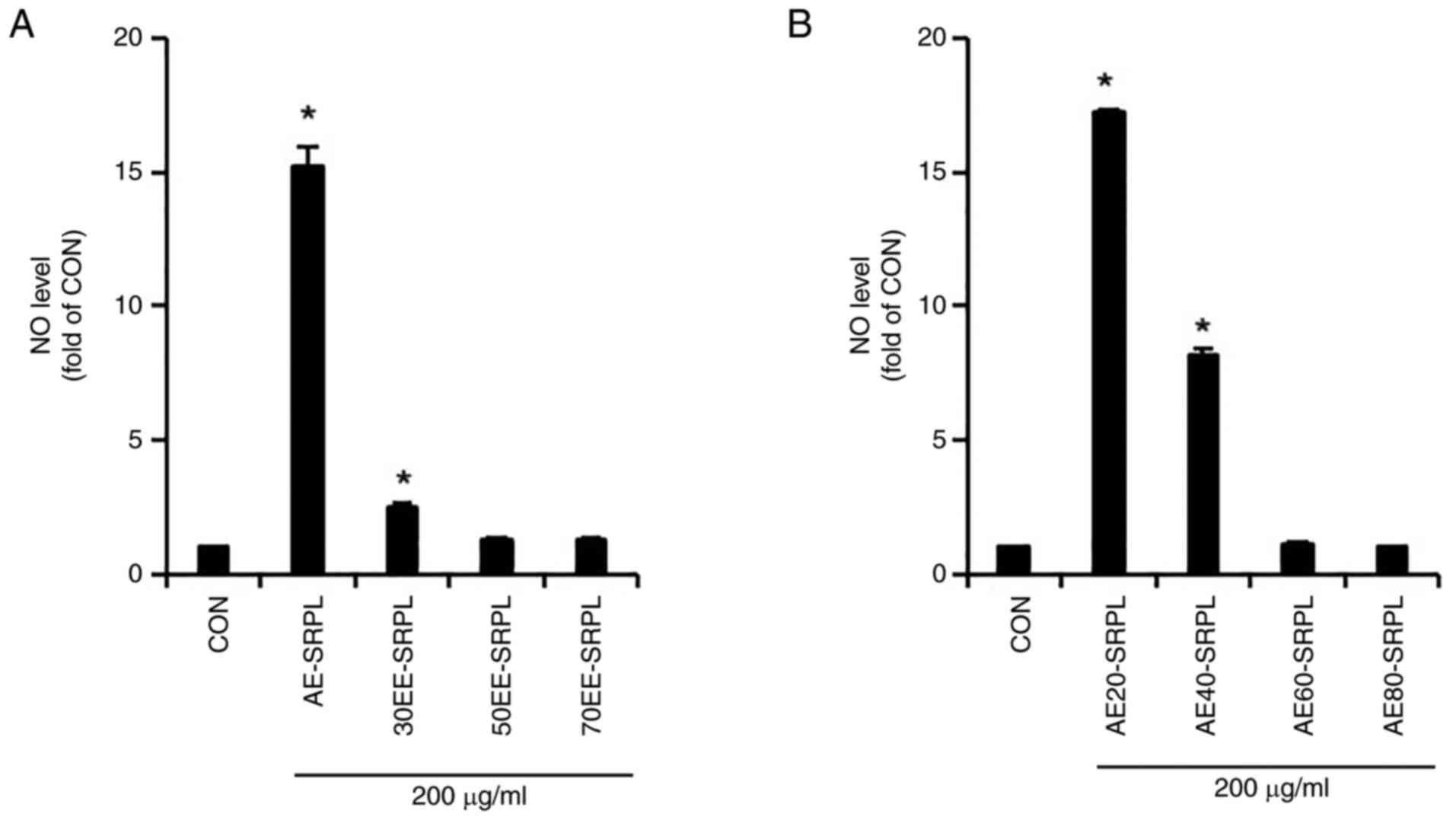 | Figure 1Comparison of NO production induction
activity based on extraction conditions of SRPL. (A) AE-SRPL,
30EE-SRPL, 50EE-SRPL or 70EE-SRPL was administered to RAW264.7
cells for 24 h. The level of NO was measured using Griess assay.
(B) AE20-SRPL, AE40-SRPL, AE60-SRPL, or AE80-SRPL was administered
to RAW264.7 cells for 24 h. The level of NO was measured using
Griess assay. *P<0.05 vs. CON. NO, nitric oxide;
AE-SRPL, aqueous extracts from SRPL; 30EE-SRPL, 30% ethanol
extracts from SRPL; 50EE-SRPL, 50% ethanol extracts from SRPL;
70EE-SRPL, 70% ethanol extracts from SRPL; AE20-SRPL, aqueous
extracts from SRPL at 20˚C; AE40-SRPL, aqueous extracts from SRPL
at 40˚C; AE60-SRPL, aqueous extracts from SRPL at 60˚C; AE80-SRPL,
aqueous extracts from SRPL at 80˚C; CON, control. |
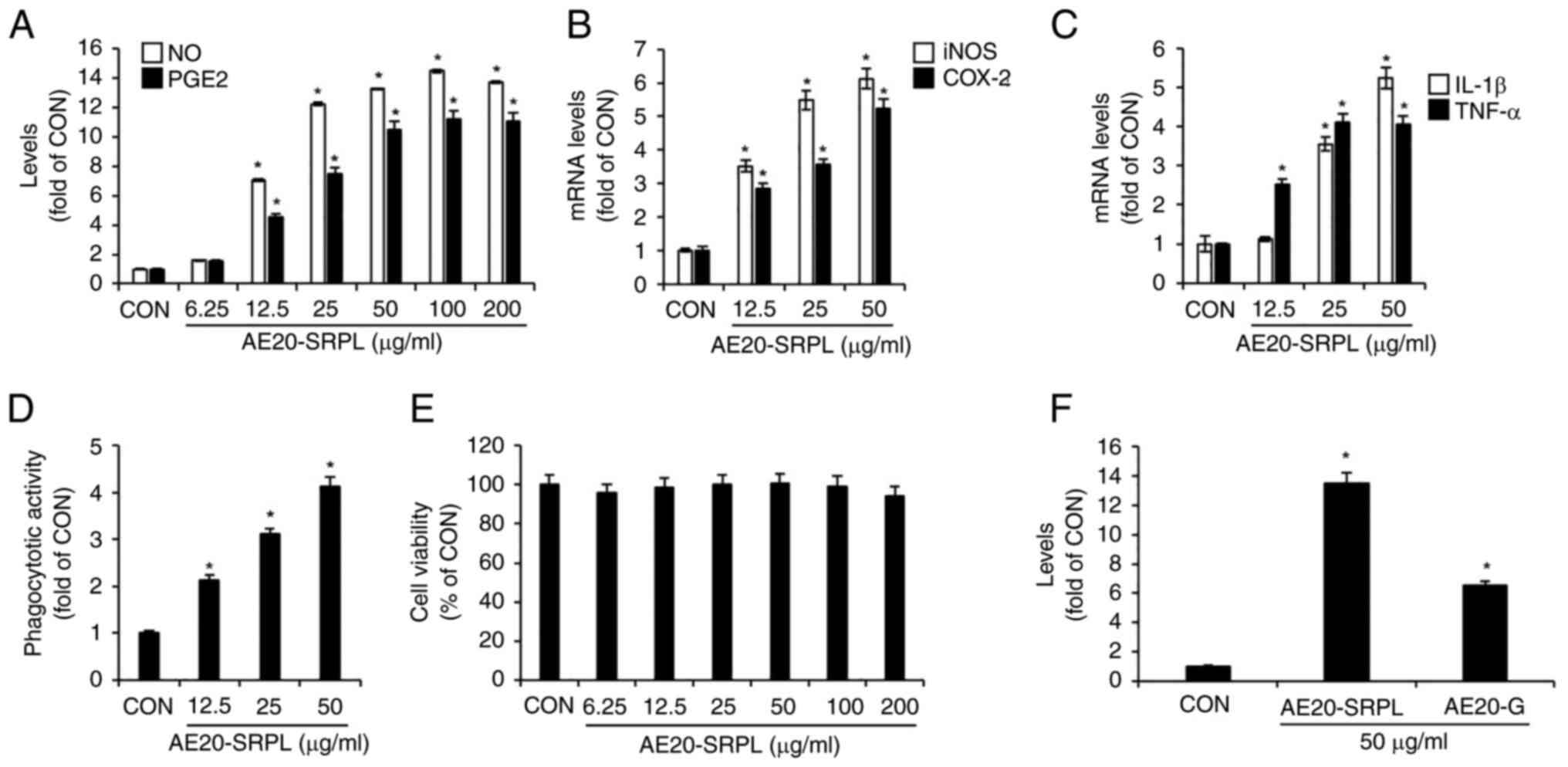 | Figure 2Effect of AE20-SRPL on macrophage
activation in RAW264.7 cells. (A) AE20-SRPL was administered to
RAW264.7 cells for 24 h. The levels of NO and PGE2 were measured
using Griess assay and ELISA kit, respectively. (B and C) AE20-SRPL
was administered to RAW264.7 cells for 24 h. The mRNA levels of
iNOS and COX-2, IL-1β and TNF-α were measured using reverse
transcription-quantitative PCR. (D) AE20-SRPL was administered to
RAW264.7 cells for 24 h. Phagocytotic activity was measured using
the neutral red uptake method. (E) AE20-SRPL was administered to
RAW264.7 cells for 24 h. Cell viability was measured using MTT
assay. (F) AE20-SRPL or AE20-G was administered to RAW264.7 cells
for 24 h. The NO level was measured using Griess assay.
*P<0.05 vs. CON. AE20-SRPL, aqueous extracts from
SRPL at 20˚C; AE20-G. aqueous extracts from ginseng at 20˚C; NO,
nitric oxide; PGE2, prostaglandin E2; iNOS, inducible nitric oxide
synthase; COX-2, cyclooxygenase-2; IL-1β, interleukin-1β; TNF-α,
tumor necrosis factor-α; CON, control. |
AE20-SRPL induces macrophage
activation in a Toll-like receptor (TLR)4-dependent manner in
RAW264.7 cells
To evaluate the contributions of TLR2 and TLR4 to
AE20-SRPL-mediated macrophage activation, the present study treated
AE20-SRPL to RAW264.7 cells in which TLR2 was inhibited by C29 or
TLR4 was inhibited by TAK-242 and then analyzed changes in the
production of NO and PGE2. The results demonstrated that inhibition
of TLR2 had no significant effect on the AE20-SRPL-mediated
increase in NO and PGE2 production (Fig. 3A). By contrast, inhibition of TLR4
markedly reduced these increases (Fig.
3A). Thus, the effect of TLR4 inhibition on AE20-SRPL-mediated
effects were investigated, specifically examining the expression of
iNOS, COX-2, IL-1β and TNF-α, as well as the activation of
phagocytic functions in macrophages. The results indicated that
TLR4 inhibition by TAK-242 significantly reduced the
AE20-SRPL-mediated expression of iNOS, COX-2, IL-1β and TNF-α
(Fig. 3B). Furthermore, this
inhibition also suppressed the activation of phagocytic functions
in macrophages induced by AE20-SRPL (Fig. 3C).
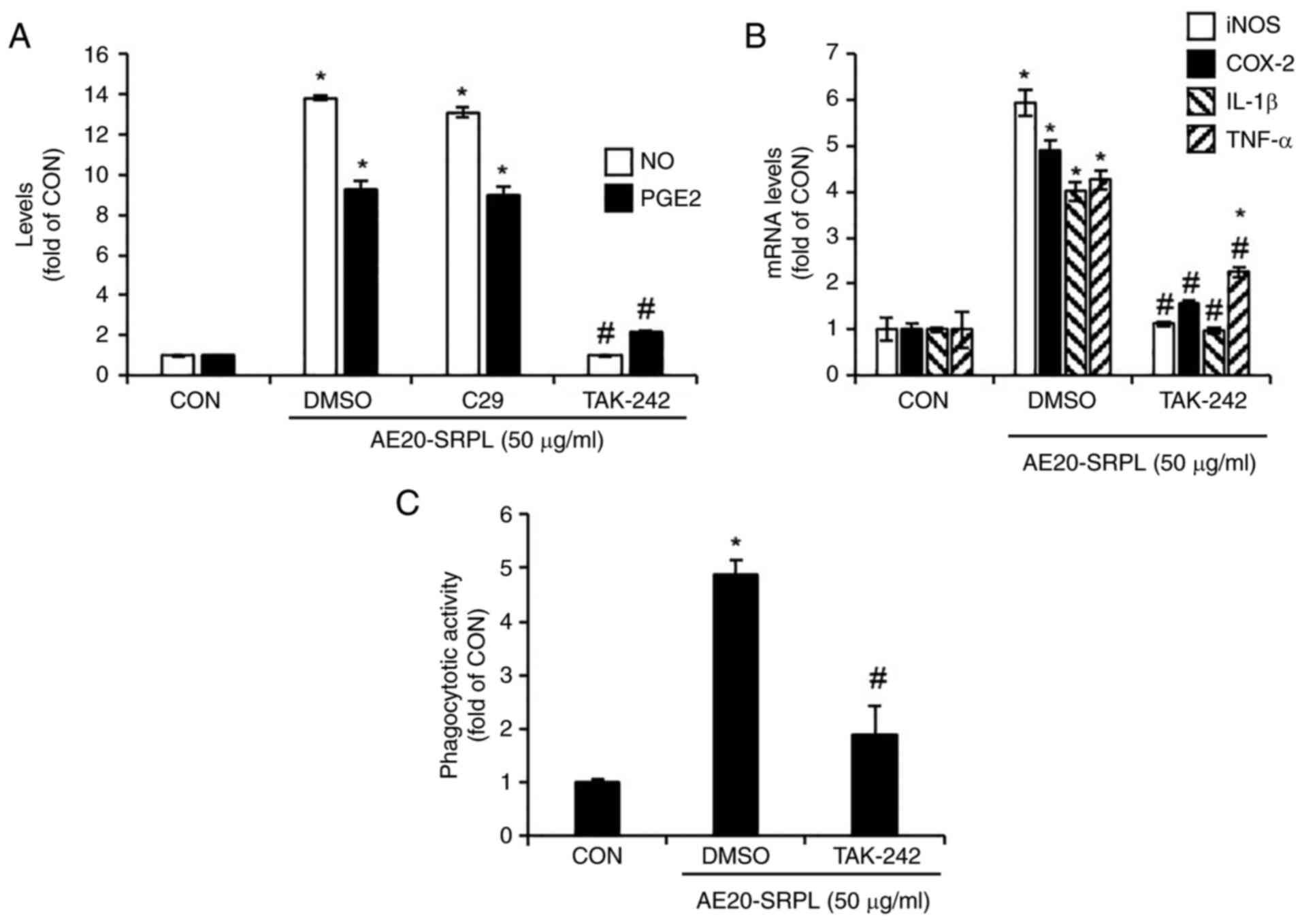 | Figure 3Effect of TLR2 and TLR4 on
AE20-SRPL-mediated activation of macrophages in RAW264.7 cells. (A)
RAW264.7 cells were pretreated with C29 (TLR2 inhibitor, 100 µM) or
TAK-242 (TLR4 inhibitor, 10 µM) for 2 h and then co-treated with
AE20-SRPL (50 µg/ml) for 24 h. The levels of NO and PGE2 were
measured using Griess assay and ELISA kit, respectively. (B)
RAW264.7 cells were pretreated with TAK-242 (TLR4 inhibitor, 10 µM)
for 2 h and then co-treated with AE20-SRPL (50 µg/ml) for 24 h. The
mRNA levels of iNOS and COX-2, IL-1β and TNF-α were measured using
reverse transcription-quantitative PCR. (C) RAW264.7 cells were
pretreated with TAK-242 (TLR4 inhibitor, 10 µM) for 2 h and then
co-treated with AE20-SRPL (50 µg/ml) for 24 h. Phagocytotic
activity was measured using the neutral red uptake method.
*P<0.05 vs. CON. #P<0.05 vs. DMSO. TLR,
Toll-like receptor; AE20-SRPL, aqueous extracts from SRPL at 20˚C;
NO, nitric oxide; PGE2, prostaglandin E2; iNOS, inducible nitric
oxide synthase; COX-2, cyclooxygenase-2; IL-1β, interleukin-1β;
TNF-α, tumor necrosis factor-α; CON, control. |
AE20-SRPL induces macrophage
activation in a JNK-dependent manner in RAW264.7 cells
To evaluate the contributions of MAPKs ERK1/2, p38
and JNK signaling to AE20-SRPL-mediated macrophage activation,
AE20-SRPL to RAW264.7 cells were treated in the absence or presence
of PD98059 (ERK1/2 inhibitor), SB203580 (p38 inhibitor) or SP600125
(JNK inhibitor) and then changes in the production of NO and PGE2
analyzed. As shown in Fig. 4A,
inhibitions of ERK1/2 by PD98059 and p38 by SB203580 had no
significant effect on the AE20-SRPL-mediated increase in NO and
PGE2 production. However, the inhibition of JNK by SP600125 blocked
these increases. Thus, it was investigated whether the inhibition
of JNK signaling pathways would also affect the AE20-SRPL-mediated
expression of iNOS, COX-2, IL-1β and TNF-α. The results
demonstrated that in RAW264.7 cells where JNK was inhibited, there
was a notable reduction in the expressions of iNOS, COX-2, IL-1β
and TNF-α by AE20-SRPL (Fig. 4B).
Furthermore, a decrease in AE20-SRPL-mediated activation of
phagocytic functions in RAW264.7 cells where JNK was individually
inhibited was observed (Fig. 4C).
To investigate whether AE20-SRPL activates JNK, the phosphorylation
of JNK in RAW264.7 cells treated with AE20-SRPL at various time
points was analyzed. As shown in Fig.
4D, the results demonstrated an enhancement in the
phosphorylation of both JNK in RAW264.7 cells following AE20-SRPL
treatment. Furthermore, it was explored whether TLR4 influences the
AE20-SRPL-mediated phosphorylation of JNK. In the absence of
TAK-242, AE20-SRPL significantly induced the phosphorylation of JNK
(Fig. 4E). However, inhibition of
TLR4 by TAK-242 resulted in a marked reduction in
AE20-SRPL-mediated phosphorylation of JNK (Fig. 4E).
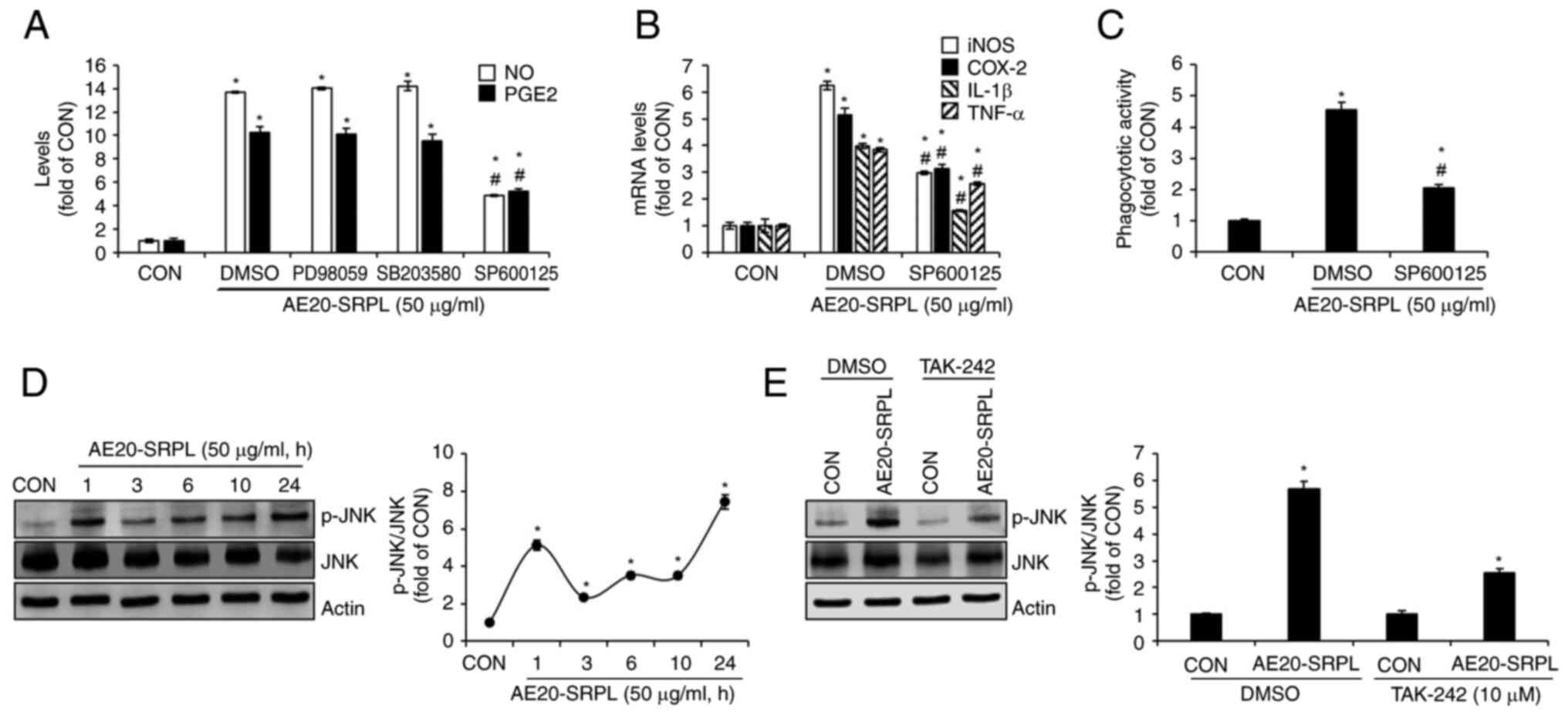 | Figure 4Effect of MAPK signaling pathways on
AE20-SRPL-mediated activation of macrophages in RAW264.7 cells. (A)
RAW264.7 cells were pretreated with PD98059 (ERK1/2 inhibitor; 20
µM), SB203580 (p38 inhibitor; 20 µM) or SP600125 (JNK inhibitor; 20
µM) and then co-treated with AE20-SRPL (50 µg/ml) for 24 h. The
levels of NO and PGE2 were measured using Griess assay and ELISA
kit, respectively. (B) RAW264.7 cells were pretreated with SP600125
(JNK inhibitor; 20 µM) and then co-treated with AE20-SRPL (50
µg/ml) for 24 h. The mRNA levels of iNOS and COX-2, IL-1β and TNF-α
were measured using reverse transcription-quantitative PCR. (C)
RAW264.7 cells were pretreated with SP600125 (JNK inhibitor; 20 µM)
and then co-treated with AE20-SRPL (50 µg/ml) for 24 h.
Phagocytotic activity was measured using the neutral red uptake
method. (D) RAW264.7 cells were treated with AE20-SRPL (50 µg/ml)
for the indicated time-points. p-JNK, JNK and Actin were measured
using western blot analysis. (E) RAW264.7 cells were pretreated
with TAK-242 (TLR4 inhibitor, 10 µM) for 2 h and then co-treated
with AE20-SRPL (50 µg/ml) for 1 h. p-JNK, JNK and Actin were
measured using western blot analysis. *P<0.05 vs.
CON. #P<0.05 vs. DMSO. AE20-SRPL, aqueous extracts
from SRPL at 20˚C; NO, nitric oxide; PGE2, prostaglandin E2; iNOS,
inducible nitric oxide synthase; COX-2, cyclooxygenase-2; IL-1β,
interleukin-1β; TNF-α, tumor necrosis factor-α; CON, control; p-,
phosphorylated; JNK, c-Jun N-terminal kinase. |
Discussion
On invasion by foreign pathogens, activated
macrophages are known to phagocytize these invaders while
concurrently secreting a diverse array of immunostimulatory factors
such as NO, PGE2, iNOS, COX-2, IL-1β and TNF-α (4). It has been reported that NO produced
by iNOS directly eliminates invading pathogens such as microbes and
viruses (13). Furthermore, NO is
known to exert a positive influence on the adaptive immune system
by promoting the differentiation and activation of T cells
(14). PGE2 synthesized by COX-2 is
also known to play a pivotal role in the human immune system,
influencing both innate and adaptive immune responses, as well as
providing critical host defenses against viral, fungal and
bacterial pathogens (15). IL-1β
contributes to the activation of macrophage phagocytic activity
against invading pathogens (16).
In addition, IL-1β has been reported to play a pivotal role as an
activator of humoral immune responses by contributing to
T-cell-dependent antibody production (17). TNF-α is recognized as a key
regulatory factor within the immune system, playing a vital role in
the immune response against various pathogens (18). TNF-α is integral to both the innate
and adaptive immune systems, with a particularly crucial function
in modulating T-cell activity within the adaptive immune response
(18). These existing reports
reflect that these immunostimulatory factors are pivotal in
modulating the efficacy and coordination of immune responses,
highlighting their integral roles in the maintenance and
enhancement of immunological functions. In immune responses, the
most critical function of macrophages is known to be their
phagocytic activity against pathogens, which initiates the innate
immune response and, in turn, orchestrates the adaptive immune
response (19). The present study
confirmed that AE20-SRPL effectively enhanced the production of
immunostimulatory factors such as NO, PGE2, iNOS, COX-2, IL-1β and
TNF-α and activated phagocytosis in RAW264.7 cells. Additionally,
it was verified that AE20-SRPL did not exhibit cytotoxic effects on
RAW264.7 cells. These findings suggested the potential of AE20-SRPL
as a natural material that can safely enhance immune function
without adverse effects.
For macrophages to produce immunostimulatory factors
through phagocytic activity, they must first recognize and become
activated by foreign pathogens via pattern recognition receptors
(PRRs) (20). This critical
recognition step facilitates the activation of macrophages,
enabling them to initiate the immune response by engulfing
pathogens and subsequently secreting a cascade of immune mediators
(20). Among PRRs, Toll-like
receptors (TLRs) are known to recognize a broad range of organisms,
including bacteria, fungi, protozoa and viruses (20). TLRs known as critical sensors for
recognizing foreign pathogens not only play an essential role in
the innate immune system but also serve as a vital bridge
connecting innate and adaptive immunity (20). Among the TLRs, TLR2 and TLR4 have
been reported to play crucial roles in recognizing foreign
pathogens and triggering the production of various
immunostimulatory factors necessary for antigen presentation
(21,22). In the present study, it was observed
that in RAW264.7 cells, the inhibition of C29 had no effect on the
production of NO and PGE2 mediated by AE20-SRPL, whereas inhibition
of TLR4 significantly reduced the generation of NO and PGE2
mediated by AE20-SRPL. Furthermore, inhibition of TLR4 also
decreased the expression of iNOS, COX-2, IL-1β and TNF-α induced by
AE20-SRPL and suppressed the activation of phagocytic activity
mediated by AE20-SRPL. These results suggested that TLR4 may play a
crucial role not only in the direct signaling mechanisms that
govern the production of immunostimulatory factors by AE20-SRPL but
also in AE20-SRPL-mediated facilitation of the phagocytic
capabilities of macrophages, which are essential for the effective
clearance of pathogens.
The MAPK pathways comprising ERK1/2, p38 and JNK are
known to be intricately linked to the activation of macrophages
(23). Thus, the present study
investigated which specific signaling pathway among ERK1/2, p38 and
JNK was utilized for AE20-SRPL-mediated macrophage activation. The
present study confirmed that the inhibition of JNK among the
ERK1/2, p38 and JNK signaling pathways reduces the production of
immunostimulatory factors and the activation of phagocytosis by
AE20-SRPL. Furthermore, AE20-SRPL activates JNK. The data suggested
that the JNK pathway is particularly critical in AE20-SRPL-mediated
macrophage activation. The immune response of macrophages to
foreign pathogens through the activation of JNK has been reported
to be dependent on TLR4(24).
Several natural agents have been reported to activate macrophages
through TLR4-dependent JNK activation (25,26).
Thus, whether TLR4 is involved in the AE20-SRPL-mediated activation
of JNK was analyzed. The results demonstrated that inhibition of
TLR4 reduces AE20-SRPL-mediated activation of JNK. These findings
indicated that the activation of JNK by AE20-SRPL may be
TLR4-dependent.
Based on the results of the present study, it can be
concluded that AE20-SRPL activates phagocytosis and enhances the
production of immunostimulatory factors through TLR4-dependent
activation of JNK in macrophages. The present study was valuable as
it identified a novel agent capable of inducing macrophage
activation for immunoenhancement and elucidates the potential
mechanism of action of this agent. However, the present study has
three limitations that necessitate further research for the
development of the immunostimulatory agent using AE20-SRPL. First,
since the present study uses an in vitro approach with
macrophages, it is imperative to conduct validation studies in
vivo to ascertain whether AE20-SRPL exhibits immunostimulatory
activity in a cyclophosphamide-induced immunosuppression C57BL/6
mice model. The second point is that the present study focused
exclusively on the activation of macrophages, which are a critical
component of the innate immune system. However, to establish robust
evidence for the immuno-enhancing activity of AE20-SRPL, it is
necessary to investigate whether AE20-SRPL also affects other types
of immune cells. Last, the present study did not conduct a detailed
analysis of the specific components of AE20-SRPL related to its
immunostimulatory activity. Therefore, it is necessary to perform
component analysis studies to identify which constituents of
AE20-SRPL are responsible for its immunostimulatory effects.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by a grant from the
National Institute of Forest Science in 2024 (project no.
FP0802-2023-01-2024) and the R&D Program for Forest Science
Technology (grant no. RS-2024-00405196) provided by the Korea
Forest Service (Korea Forestry Promotion Institute, Seoul,
Korea).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HJC, GHP, JWC, SJP, JHH, SHL, HYK and MYC performed
cell-based experiments and analyzed the data. HJC and GHP wrote the
manuscript. JBJ designed the experiments and wrote and edited the
manuscript. HJC, GHP, JWC, SJP, JHH, SHL, HYK, MYC and JBJ confirm
the authenticity of all the raw data. All the authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hirayama D, Iida T and Nakase H: The
phagocytic function of macrophage-enforcing innate immunity and
tissue homeostasis. Int J Mol Sci. 19(92)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Duque GA and Descoteaux A: Macrophage
cytokines: Involvement in immunity and infectious diseases. Front
Immunol. 5(491)2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sieweke MH and Allen JR: Beyond stem
cells: self-renewal of differentiated macrophages. Science.
342(1242974)2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hume DA: The mononuclear phagocyte system.
Curr Opin Immunol. 18:49–53. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Muntjewerff EM, Meesters LD and van den
Bogaart G: Antigen cross-presentation by macrophages. Front
Immunol. 11(1276)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gordon S and Mantovani A: Diversity and
plasticity of mononuclear phagocytes. Eur J Immunol. 41:2470–2472.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shin MS, Hwang SH, Yoon TJ, Kim SH and
Shin KS: Polysaccharides from ginseng leaves inhibit tumor
metastasis via macrophage and NK cell activation. Int J Biol
Macromol. 103:1327–1333. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tabarsa M, Jafari A, You S and Cao R:
Immunostimulatory effects of a polysaccharide from Pimpinella
anisum seeds on RAW264.7 and NK-92 cells. Int J Biol Macromol.
213:546–554. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang A, Yang X, Li Q, Yang Y, Zhao G,
Wang B and Wu D: Immunostimulatory activity of water-extractable
polysaccharides from Cistanche deserticola as a plant adjuvant in
vitro and in vivo. PLoS One. 13(e0191356)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lim HI: Seed dormancy and germination
characteristics of endemic elder species (Sambucus racemose subsp.
pendula) and common elder species (S. williamsii) in Korea. J For
Environ Sci. 38:284–289. 2022.
|
|
11
|
Mocanu ML and Amariei S: Elderberries-A
source of bioactive compounds with antiviral action. Plants
(Basel). 11(740)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bogdan C: Nitric oxide synthase in innate
and adaptive immunity: An update. Trends Immunol. 36:161–178.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
García-Ortiz A and Serrador JM: Nitric
oxide signaling in T cell-mediated immunity. Trends Mol Med.
24:412–427. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Martínez-Colón GJ and Moore BB:
Prostaglandin E2 as a regulator of immunity to
pathogens. Pharmacol Ther. 185:135–146. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Netea MG, Simon A, van de Veerdonk F,
Kullberg BJ, van der Meer JWM and Joosten LAB: IL-1β processing in
host defense: Beyond the inflammasomes. PLoS Pathog.
6(e1000661)2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Nakae S, Asano M, Horai R and Iwakura Y:
Interleukin-1beta, but not interleukin-1α, is required for
T-cell-dependent antibody production. Immunology. 104:402–409.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vielhauer V and Mayadas TN: Functions of
TNF and its receptors in renal disease: Distinct roles in
inflammatory tissue injury and immune regulation. Semin Nephrol.
27:286–308. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Aderem A and Underhill DM: Mechanisms of
phagocytosis in macrophages. Ann Rev Immunol. 17:593–623.
1999.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhou L, Cao X, Fang J, Li Y and Fan M:
Macrophages polarization is mediated by the combination of PRR
ligands and distinct inflammatory cytokines. Int J Clin Exp Pathol.
8:10964–10974. 2015.PubMed/NCBI
|
|
21
|
Beutler B, Jiang Z, Georgel P, Crozat K,
Croker B, Rutschmann S, Du X and Hoebe K: Genetic analysis of host
resistance: Toll-like receptor signaling and immunity at large. Ann
Rev Immunol. 24:353–389. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Takeda K and Akira S: Toll-like receptors
in innate immunity. Int Immunol. 17:1–4. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ren D, Lin D, Alim A, Zheng Q and Yang X:
Chemical characterization of a novel polysaccharide ASKP-1 from
Artemisia sphaerocephala Krasch seed and its macrophage activation
via MAPK, PI3k/Akt and NF-κB signaling pathways in RAW264.7 cells.
Food Funct. 8:1299–1312. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Swanson L, Katkar GD, Tam J, Pranadinata
RF, Chareddy Y, Coates J, Anandachar MS, Castillo V, Olson J, Nizet
V, et al: TLR4 signaling and macrophage inflammatory responses are
dampened by GIV/Girdin. Proc Natl Acad Sci USA. 117:26895–26906.
2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Um Y, Eo HJ, Kim HJ, Kim K, Jeon KS and
Jeong JB: Wild simulated ginseng activates mouse macrophage,
RAW264.7 cells through TRL2/4-dependent activation of MAPK, NF-κB
and PI3K/AKT pathways. J Ethnopharmacol. 263(113218)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xie XD, Tang M, Yi SL, He Y, Chen SY, Zhao
Y, Chen Q, Cao MX, Yu ML, Wei YY, et al: Polysaccharide of
Asparagus cochinchinensis (Lour.) Merr regulates macrophage immune
response and epigenetic memory through TLR4-JNK/p38/ERK signaling
pathway and histone modification. Phytomedicine.
124(155294)2024.PubMed/NCBI View Article : Google Scholar
|


















