Introduction
The ‘Guidelines for Severe Community-Acquired
Pneumonia (CAP)’ released in the United States in 2019 indicate
that the prevalence of atypical pathogens, especially Legionella
pneumophila, has significantly increased in previous years
(1,2). Legionella pneumophila is a
common atypical pathogen that causes pneumonia and is the leading
cause of hospital admission in ~2-15% of patients with CAP
(3,4). Legionella gormanii is a
gram-negative bacterium belonging to the genus Legionella
that is closely related to Legionnaires' disease in humans. The
main clinical symptoms include fever, cough, sore throat, runny
nose, limb or joint pain, headache, vomiting and diarrhoea. The
main clinical symptoms are not specific, and the prevalence of
influenza A subtype (H1N1) is similar to that of Legionella
pneumophila (5). Older
individuals with Legionella and H1N1 influenza virus
coinfection are prone to misdiagnosing the influenza A subtype
(H1N1) virus, which can result in a delay in administering
appropriate antibiotic treatment. Importantly, this delay may be
associated with worsening morbidity and mortality (6).
Traditional Legionella culture methods are
time-consuming, and the cultures are susceptible to contamination
(7). Clinically, the diagnosis of
Legionella infection often relies on the detection of urine
antigens and anti-Legionella antibodies (8). However, the appearance of
anti-Legionella antibodies is relatively delayed, typically
occurring 3 weeks after onset. The clinical specificity of their
detection is very poor, and the currently widely used urine antigen
test is available only for detecting the Legionella
pneumophila serogroup (9). To
meet the clinical requirements for the rapid diagnosis and timely
treatment of Legionella infection, metagenomic
next-generation sequencing (mNGS) is a new tool that can quickly
and accurately identify potential pathogens. mNGS was previously
highlighted as the most promising method for comprehensively
diagnosing infections, particularly severe pneumonia, in the
intensive care unit (ICU) (10). In
our clinical laboratory, a mNGS platform was build for the
diagnosis of infectious disease. The process included nucleic acid
extraction, library construction, sequencing, bioinformatics
analysis and result interpretation. QIAamp® kits were
used nucleic acid extraction in clinical samples. Library
construction was performed with the Nextera XT DNA Library Prep Kit
(Illumina, Inc.). Sequencing on the Illumina Nextseq CN500 used
SE-75 or SE-50 protocols, generating ~20 million reads/sample.
Low-quality reads and human sequences were filtered out, followed
by microbial database alignment for species identification. Result
interpretation guidelines are detailed in the authors' previous
studies (11,12). The aim of the present study was to
explore the clinical features, diagnosis and treatment of a severe
pneumonia patient with co-infection of Legionella gormanii
and influenza A subtype (H1N1) virus. Our mNGS technology was
utilized to promptly and accurately diagnose Legionella
gormanii, providing patients with the opportunity for early
treatment.
Case report
A 79-year-old man with a history of high blood
pressure, diabetes, cerebral infarction and tuberculosis was
admitted to the ICU on February 28, 2023, at the First Affiliated
Hospital, Zhejiang University School of Medicine (day 1), due to
repeated fever, cough and sputum over the previous 10 days. At that
time, he was diagnosed with CAP and was administered
piperacillin-tazobactam (4.5 g q8h) as empirical therapy. The
patient's body temperature improved (specific details are unknown),
but there was no significant improvement in the symptoms of cough
or sputum production. The patient denied travel history, exposure
to any cooling systems or other special man-made water system
exposure. In addition, blood gas analysis and whole blood lactate
measurements revealed the following results: The partial pressure
of carbon dioxide was 41.3 mmHg, and the partial pressure of oxygen
was 53.8 mmHg. Maintaining adequate oxygenation with nasal
high-flow oxygen (maximum oxygen concentration of 85%) remains
challenging. The patient exhibited respiratory failure and severe
pneumonia and required endotracheal intubation and
ventilator-assisted ventilation (PCV-A/C, FiO2: 35%,
SpO2 90-95%) (Fig. 1A).
The pneumonia severity index was >130 points.
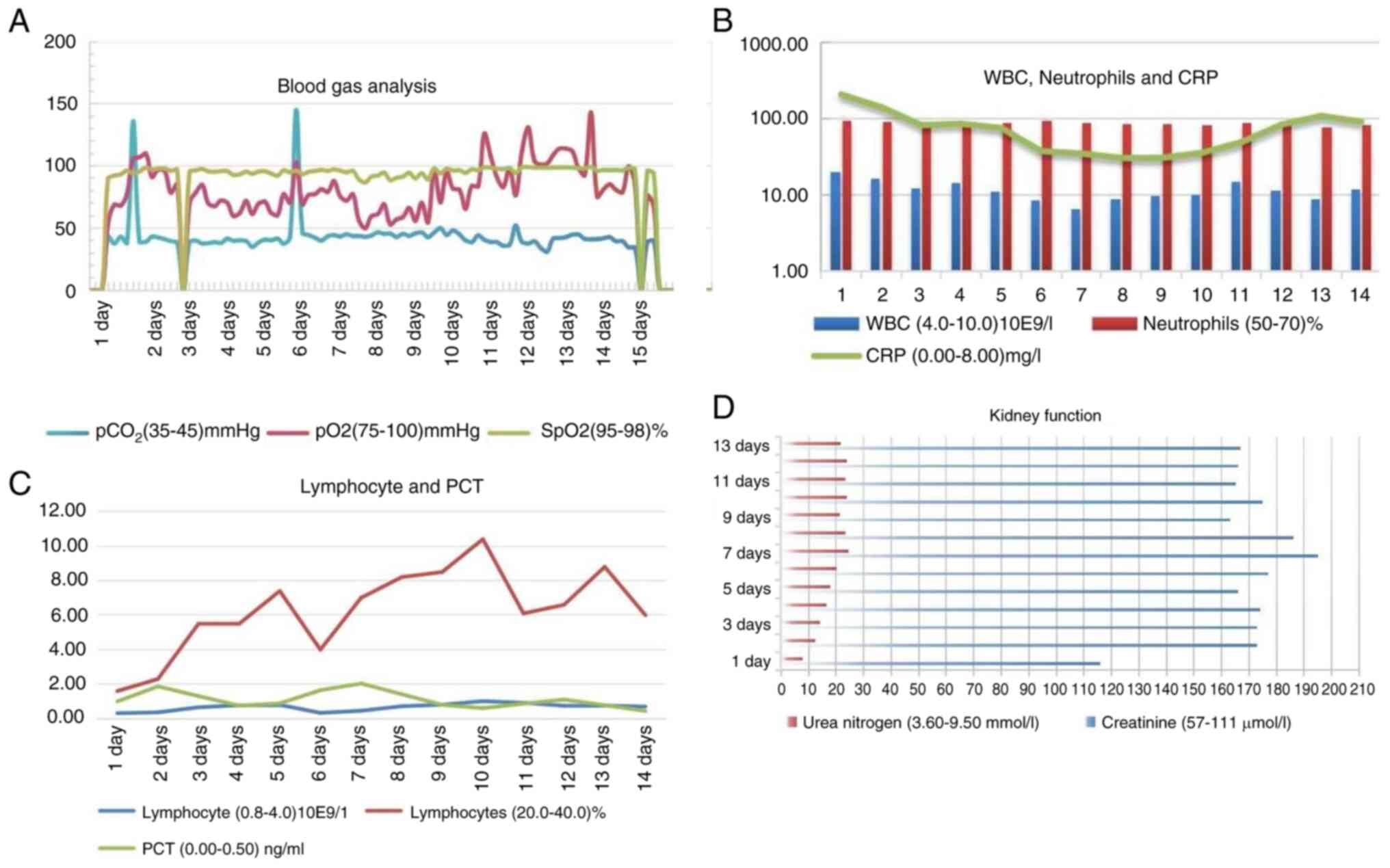 | Figure 1Regularity of inflammatory indicators
in patients. (A) Changes in the pCO2, pO2 and
SpO2 in the patient's arterial blood before and after
receiving ventilator therapy. (B) After treatment, the CRP levels
decreased significantly, from 209.34 to a minimum of 30.52 mg/l.
(C) Changes in peripheral blood lymphocytes and PCT levels after
treatment: PCT decreased significantly, while the lymphocyte count
remained low. (D) Urea nitrogen levels consistently exceeded the
upper limit of detection, ranging from 7.93 to 24.5 mmol/l, while
creatinine levels fluctuated between 116 and 195 µmol/l. WBC, white
blood cell; CRP, C-reactive protein; PCT, procalcitonin;
pCO2, arterial partial pressure of carbon dioxide;
pO2, arterial partial pressure of oxygen;
SpO2, arterial oxygen saturation. |
Physical examination revealed a temperature of
36.1˚C, a pulse of 108 beats/min, 25 breaths/min, and a blood
pressure of 149/110 mmHg. Laboratory investigations revealed a
white blood cell count of 19.82x109/l (4.0-10.0), with a
neutrophil percentage of 92.8% (50.0-70.0%), a lymphocyte count of
0.32x109/l (0.8-4.0/l), a monocyte count of
1.07x109/l (0.12-1.00/l), a procalcitonin (PCT)
concentration of 1.00 ng/ml (0.00-0.05 ng/ml) and a C-reactive
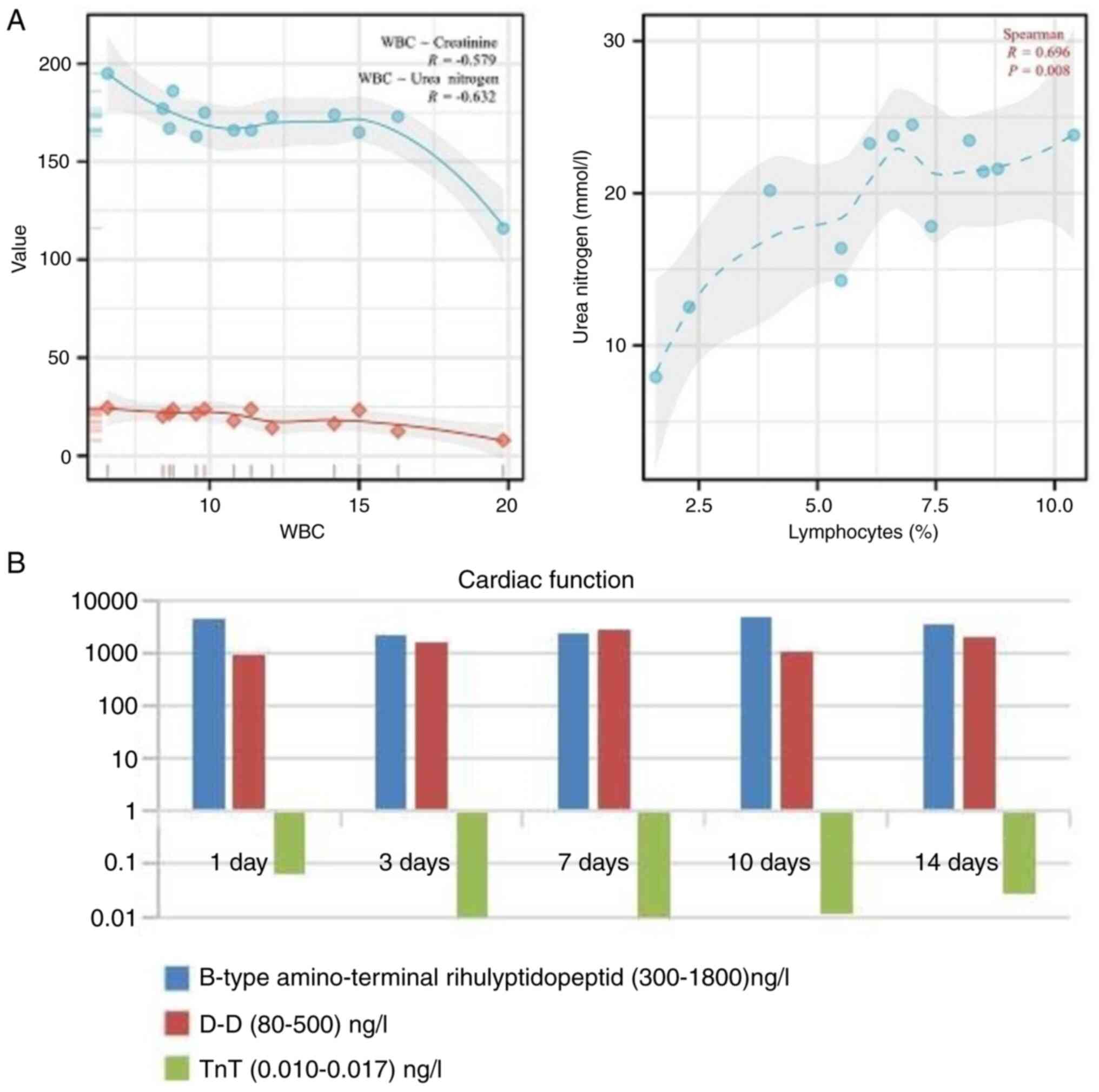
protein (CRP) level of 209.34 mg/l (0.00-8.00 mg/l) (Fig. 1B and C). The serum creatinine concentration was
116 µmol/l (57-111 µmol/l), and the urea nitrogen concentration was
7.53 mmol/l (3.60-9.50 mmol/l) (Fig.
1D). The indicators of renal function appeared to be within
normal limits. The B-type amino-terminal natriuretic peptide level
was 4,600 ng/l (300-1,800 ng/l), the D-D level was 917 ng/l (80-500
ng/l) and the TnT level was 0.067 ng/l (0.010-0.017 ng/l) (Fig. 2A and B). Cardiac function indicates significant
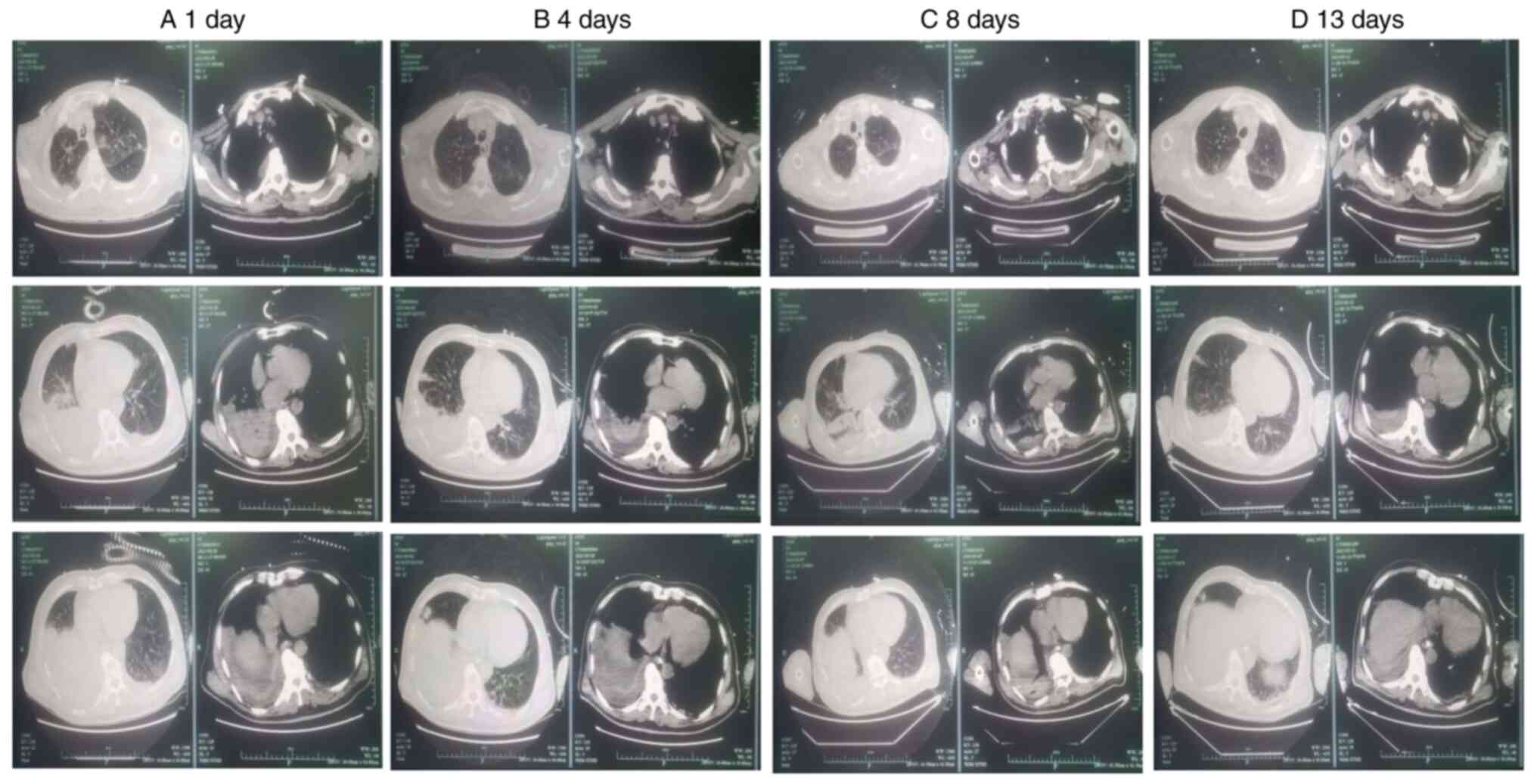
functional deficiencies. Chest computed tomography (CT) performed
on February 28 (day 1) revealed two signs of pneumonia:
consolidation in the lower lobe of the right lung and a significant
amount of fluid in the chest cavity on both sides (Fig. 3A). Moreover, the bronchoalveolar
lavage fluid (BALF) mNGS were conducted to identify the potential
pathogen.
On the second day, the BALF mNGS results revealed
Legionella gormanii and H1N1 influenza infection on March 1
(Fig. 4). DNA mNGS detected 665
sequences that could be mapped to Legionella gormanii out of
a total of 16,851,224 sequences, with a coverage of 0.892% and
13.856%, respectively (Fig. 4A).
RNA mNGS detected 1,458 sequences that could be mapped to H1N1
influenza virus in a total of 6,398,711 sequences, with a coverage
of 89.246% (Fig. 4B). The patient
was diagnosed with community-acquired pneumonia caused by
Legionella gormanii and coinfection with the H1N1 influenza
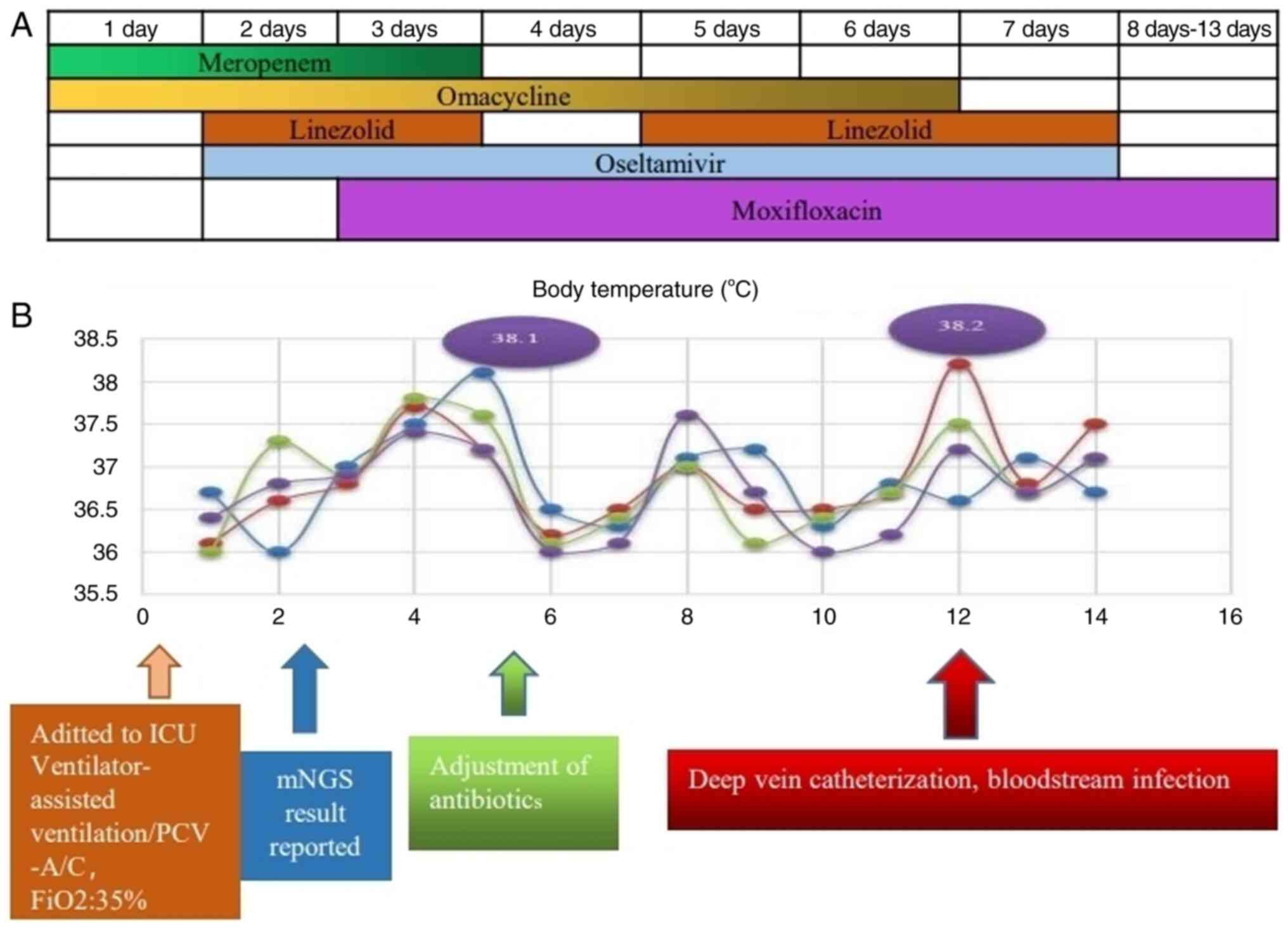
virus. The patient received timely symptomatic treatment, which
included an intravenous drip of linezolid (0.6 g Q12H) and oral
oseltamivir (75 mg Q12H). When the patient's temperature continued
to rise, an intravenous drip of moxifloxacin (400 mg QD) and other
treatments were administered (Fig.
5A).
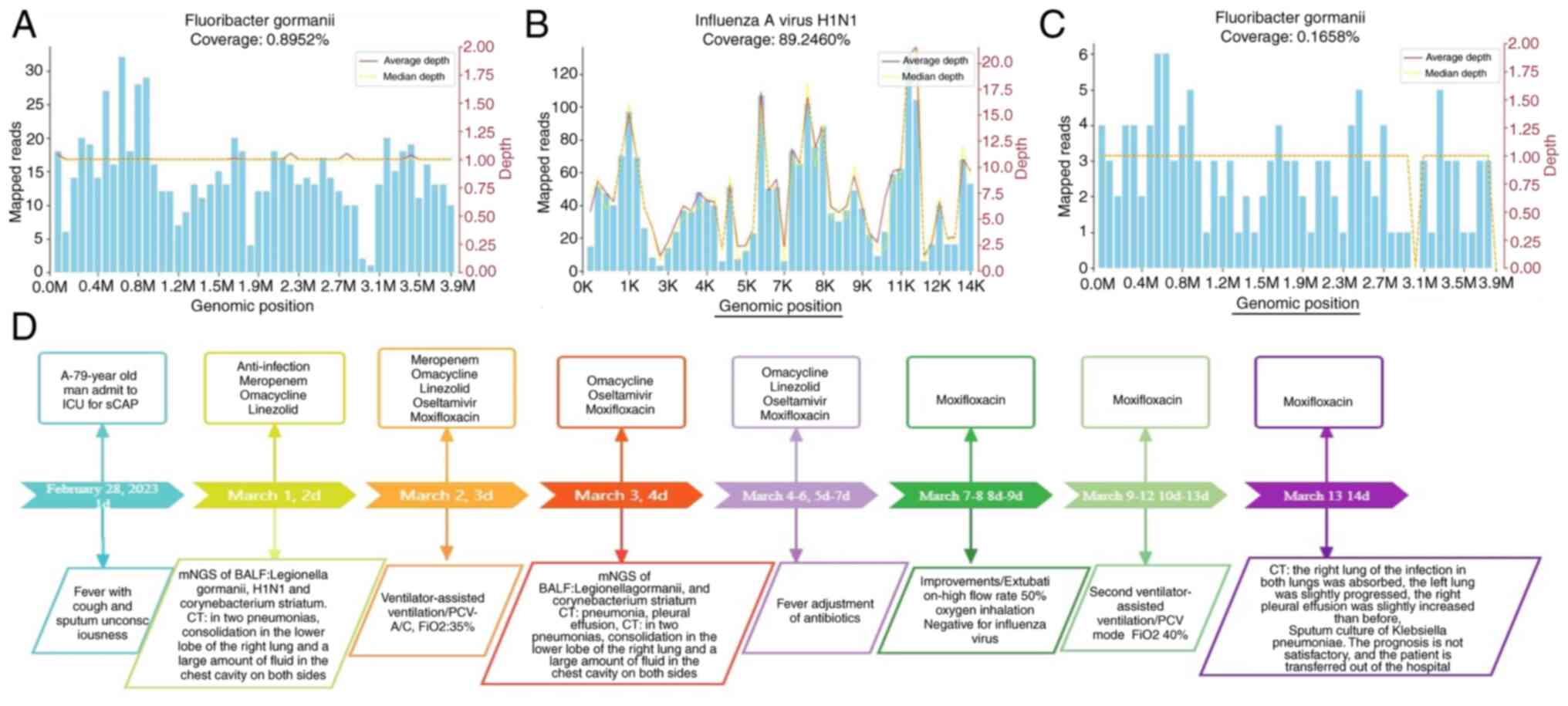 | Figure 4Sequence characteristics of
Legionella gormanii H1N1 detected by mNGS during treatment.
(A) In a total of 16,851,224 sequences, DNA mNGS detected 665
sequences that could be mapped to Legionella gormanii; the
coverage was 0.892% and 13.856% (green columnar section). (B) RNA
mNGS detected 1,458 sequences mapped to H1N1 influenza virus in a
total of 6,398,711 sequences, and the coverage was 89.246% (green
columnar section). (C) After 5 days of treatment, the number of raw
reads of Legionella gormanii in BALF was 112, and the
coverage was 0.165% (green column). (D) A brief review of the
medical history and treatment of this 79-year-old patient. mNGS,
next-generation sequencing; BALF, bronchoalveolar lavage fluid;
ICU, intensive care unit. |
After treatment on the fourth day, the patient's
body temperature remained normal (Fig.
5B), and his overall condition improved significantly. The
levels of inflammatory indicators, such as PCT, CRP and white blood
cells, decreased significantly (Fig.
1B and C). CT imaging revealed
two signs of pneumonia: Consolidation in the lower lobe of the
right lung, which had partially resolved, and a small amount of
fluid in the pleural cavity on both sides (Fig. 3B). Tests for β-(1,3)-glucan
(BD) and galactomannan were negative. Linezolid treatment was
discontinued on the 4th day, and the patient developed a fever on
the fifth day, reaching 38.2˚C, but the CRP level did not
significantly increase. After 1 week of treatment, the number of
raw reads of Legionella gormanii in BALF was 112, with a
coverage of 0.165%. The virus test result was retested, and the
results were negative according to reverse transcription (RT)-PCR.
For empirical anti-infective therapy, these results indicated that
the treatment was effective, and the chest CT changes were
consistent (Figs. 4C and 3C). After antibiotic treatment, the
indicators of kidney function damage significantly increased
(creatinine 195 µmol/l, urea nitrogen 24.5 mmol/l), and the
indicators of heart damage also significantly increased (B-type
amino-terminal rihulyptidopeptide 2,450 ng/l, D-D 2,870 ng/l)
(Figs. 1D and 2B). The organ function was not promising,
and the correlation analysis revealed a significant relationship
between white blood cell count, creatinine and urea nitrogen,
showing negative correlations (R=-0.596, R=-0.632). Additionally,
there was a positive correlation between lymphocyte percentage and
urea nitrogen (R=0.696) (Fig. 2A).
The impact of drug side effects cannot be disregarded when the
impairment of kidney function is associated with the virus, as
heart and lung function are closely interconnected. Furthermore,
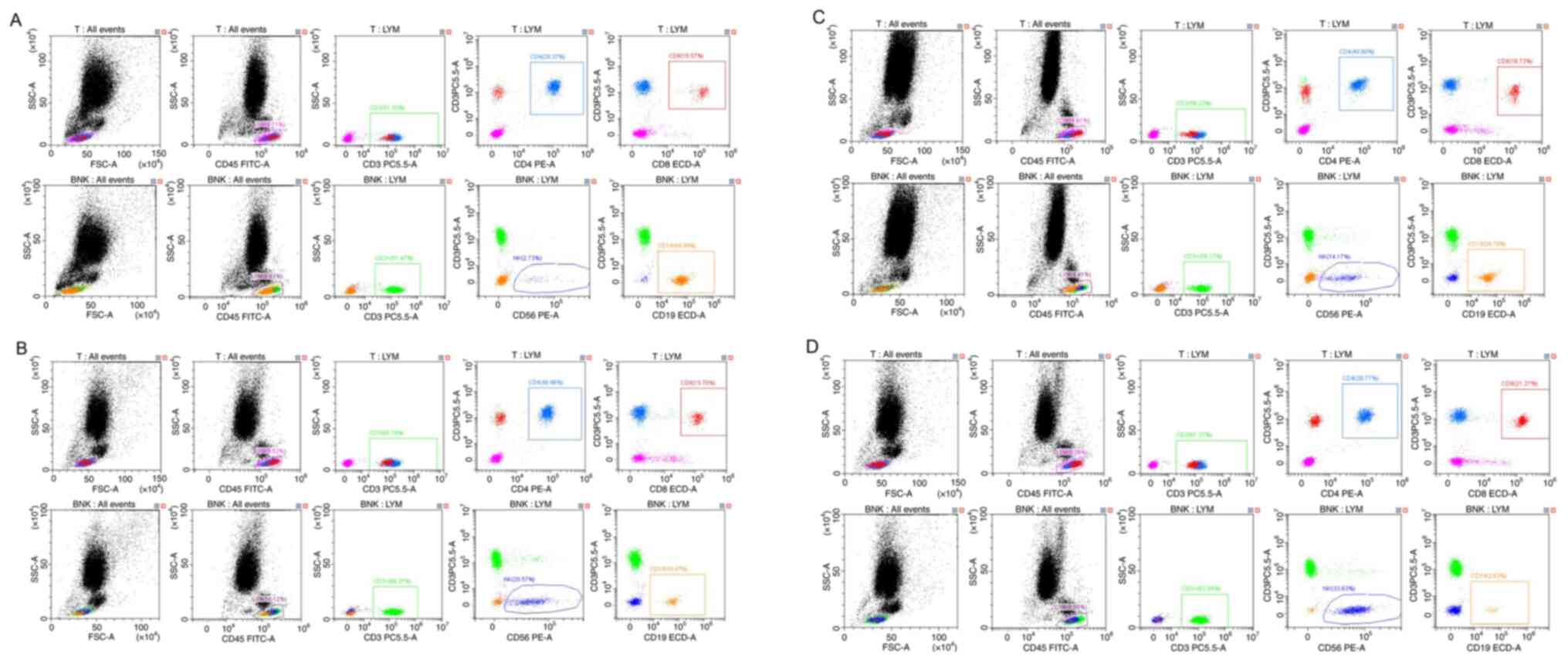
The FACSCanto flow cytometer (Becton Dickinson and Company) and a
set of six-colour fluorescently labelled antibodies, including i)
ISG1, ii) fluorescein isothiocyanate (FITC)/IgG1, iii) Phycocyanin
(PE), iv) CIM FITC/CD8-PE, v) CD3-FITC/CD16+56 PE and
CD19-ECD (Becton, Dickinson and Company) were utilized. The
Lymphocyte Subsets Test Kit (Becton, Dickinson and Company; cat.
no. 662967) was utilized to measure the ratio of T cell subsets
(CD3+ CD4+, CD3+ CD8+),
B cells (CD3-CD19+) and the percentage of natural killer
(NK) cells (CD3-CD16+ CD56+) in peripheral
blood, providing relative and absolute values of the detected
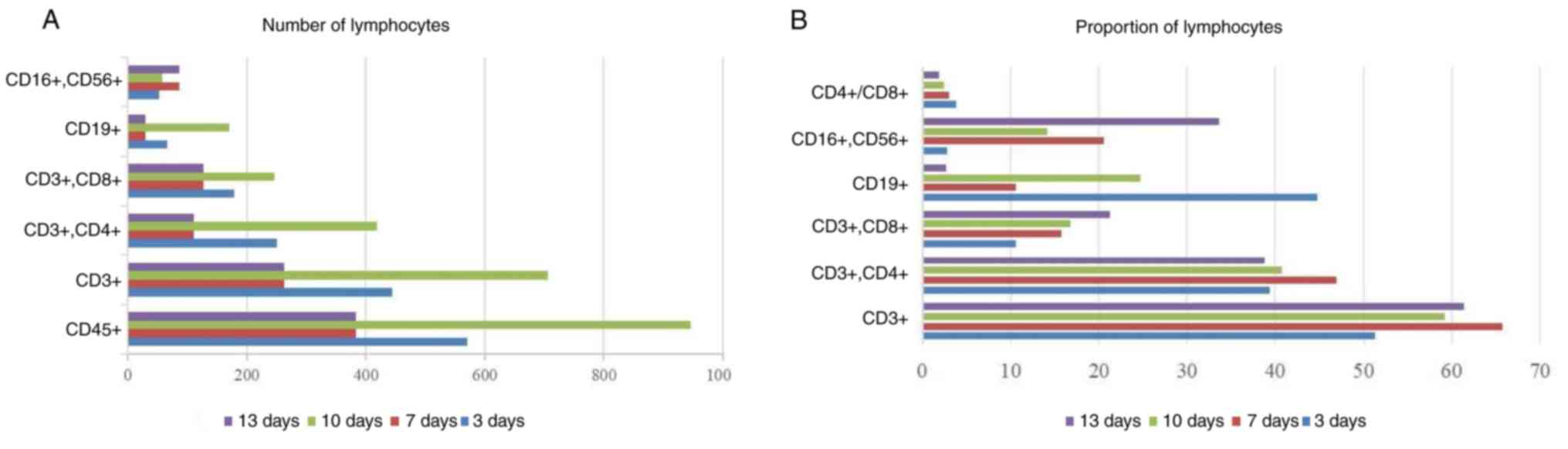
immune cells. The absolute number of CD45 decreased from 570 to
383, CD3 decreased from 444 to 263, the relative number varied from
51.33 to 61.37%, the absolute value of CD19 decreased from 66 to
29, and the relative value decreased from 44.8 to 2.63%. However,
the absolute and relative counts of CD16+ and
CD56+ both increased, from 86 and 2.73 to 33.63% from
the original 52. The decrease in T and B cells reflects the poor
immune function of the patient, and the increase in NK cells was
caused by the elimination effect of Legionella and viruses
on these foreign pathogens (Figs.
6A and B and 7A-D).
After 13 days of treatment, the CRP index increased.
CT imaging revealed significant inflammatory infiltrative changes
in the lungs. However, the function of the left lung had slightly
deteriorated. The severity of the right pleural effusion also
slightly increased, and sputum culture revealed Klebsiella
pneumoniae (Fig. 3D). Spearman
correlation analysis was conducted using SPSS 20 software (IBM
Corp.). There was no significant improvement in cardiopulmonary
function (Fig. 2B). The patient's
condition remained unstable due to the lack of significant
improvement in their immune system, compounded by the underlying
primary disease. In this scenario, new pathogens associated with
nosocomial infections and multiple antibiotic-resistant strains
have emerged, leading to no apparent improvement following
treatment. Consequently, the patient's family requested discharge.
The treatment process is illustrated in Fig. 4D.
Discussion
To understand the clinical features, diagnosis and
treatment of Legionella gormanii in conjunction with
influenza A subtype (H1N1) virus, which causes a high-risk,
low-epidemic infectious disease, the present case report introduced
a patient with Legionella gormanii and H1N1 influenza virus
coinfection, which led to severe pneumonia. mNGS technology was
utilized to promptly and accurately diagnose Legionella
gormanii, providing patients with the opportunity for early
treatment.
Legionella is a significant cause of
community-acquired pneumonia, with ~90% of reported cases
attributed to Legionella pneumophila, 79% of which are
caused by the Legionella pneumophila serogroup (13). Human infection with Legionella
pneumophila primarily occurs through the inhalation of aerosols
containing pathogens (14).
However, the symptoms of H1N1 influenza are constantly evolving,
especially in older individuals with underlying medical conditions.
Older individuals with Legionella and H1N1 influenza virus
coinfection are prone to misdiagnosis, which can delay the
administration of antibiotic treatment.
Previous cases were reviewed of misdiagnosis and
missed diagnoses of Legionella pneumonia (Table I) (12). According to the relevant literature,
the patient tested positive for the Legionella antigen in
his urine. However, there were also instances of false positive
results in urine tests, and the testing process was relatively
time-consuming. There are several crucial factors to consider in
the misdiagnosis of Legionella pneumonia. First, the
clinical manifestations of Legionella pneumoniae infection
are non-specific, and the diagnosis is based on laboratory testing
for pathogens. Second, molecular diagnostic techniques, with a
sensitivity of 70-80% and high specificity of 99-100%, have
revealed a high prevalence of respiratory viruses in cases of
atypical bacterial infections. Compared with 16S rRNA gene
sequencing (Cloning library sequencing; Applied Biosystems; Thermo
Fisher Scientific, Inc.), mNGS offers greater classification
resolution and has been utilized in pneumonia diagnosis, outbreak
tracking, infection control monitoring and pathogen detection
(15).
 | Table IA review of clinical information on
Legionella gormanii combination influenza A subtype (H1N1) virus
syndrome cases reported in recent years. |
Table I
A review of clinical information on
Legionella gormanii combination influenza A subtype (H1N1) virus
syndrome cases reported in recent years.
| Case/Sex/Age | Symptoms and
Inducement | Clinical feature | Pathogen (detection
method) | Medication | Length of stay | Assisted
ventilation | Outcome | References |
|---|
| The present
case/Male/79 | Repeated fever with
cough and sputum during the previous 10-day | Respiratory failure.
Severe pneumonia. High blood pressure. Diabetes | mNGS of BALF:
Legionella gormanii H1N1 | Meropenem Omacycline
Linezolid Oseltamivir Moxifloxacin | 14 days | Trachea cannula | Transfer to hospital
for rehabilitation | The present
study |
| Case1/Male/59 | Hot/cold sweats and
high fever shower in the work's changing rooms were not widely
used | Pulmonary infiltrates
atrial flutter with rapid ventricular response | Urinary legionella
antigen: Legionella gormanii RT-PCR: NH1N1 | Oseltamivir
rifampicin clarithromycin ciprofloxacin | 14 days | | Live | Schofield et
al, 2010(16) |
| Case 2 | Sore throat without
obvious | Exacerbations of
chronic bronchitis, asthma and congestive heart failure | Urinary legionella
antigen: Legionella gormanii RT-PCR: NH1N1 | Oseltamivir other
antibiotics | | | Live | Burk et al
2010(5) |
| Case 3 | | Pneumonia after
returning from a 1-week travel abroad | Legionella serology
(single titer): Legionella gormanii RT-PCR: NH1N1 | Oseltamivir other
antibiotics | | | Live | Caterina et
al, 2010(12) |
Severe community-acquired pneumonia was definitively
diagnosed, which was caused by Legionella gormanii in
combination with influenza A subtype (H1N1) in an immunocompetent
patient, as detected by mNGS. Fluoroquinolones or macrolides are
considered first-line options for patients with Legionella
pneumonia, while combination therapy is recommended for
critically ill or immunocompromised patients. The initial treatment
for the H1N1 virus was 75 mg of oseltamivir twice daily, and the
patient was relocated to an isolated room in accordance with the
hospital's infection control policy. After 10 days of antibiotic
treatment, the patient's overall condition significantly improved,
and the levels of inflammatory biomarkers decreased. CT imaging
twice demonstrated that the pleural effusion had significantly
decreased, indicating absorption, and the RT-PCR test result was
negative. On days 11-14, the patient had a fever and elevated
levels of the inflammatory marker CRP. Kidney function damage
significantly increased (Fig. 1D
and F). CT imaging of the chest
revealed enlarged pneumonia opacities on both sides and increased
pleural effusions on both sides, while sputum culture indicated the
presence of drug-resistant Klebsiella pneumoniae. The
occurrence of Klebsiella pneumoniae is mainly due to
endogenous infection in the hospital, primarily through
self-contact transmission of pulmonary gram pathogens within the
patient's body (16,17). These changes predict worsening of
the condition and a poor prognosis.
The worsening of the patient's condition is likely
due to the presence of multiple underlying diseases, such as
hypertension and diabetes. Additionally, the patient was
definitively diagnosed with Legionella pneumonia (6). The appropriate treatment requires a
significant amount of time, which can delay the recovery process
and cause the patient to miss the optimal treatment window. Third,
the patient's inflammatory markers did not decrease to normal
levels, and her immune function continued to deteriorate. Fourth,
there was no significant recovery from severe cardiac function
injury, as indicated by high levels of B-type amino-terminal
rihulyptidopeptidase, and the elevated D-dimer suggested that the
patient's peripheral blood circulation had not improved (18).
In summary, the present study aimed to investigate
the clinical characteristics, diagnosis, and management of
Legionella gormanii in conjunction with the influenza A
subtype (H1N1) virus. This combination results in a high-risk,
low-epidemic infectious disease. MNGS technology was utilized to
promptly and accurately diagnose Legionella gormanii.
Patients who respond to antibiotic and antiviral treatments. mNGS
may be a high-resolution and sensitive assay for the diagnosis and
surveillance of Legionella infection. Further research and
exploration are still needed to understand the pathogenic mechanism
of Legionella and to evaluate the effectiveness of
antibiotics.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Zhejiang Provincial
Natural Science Foundation (grant no. LY23H200001).
Availability of data and materials
The data generated in the present study may be found
in the in the National Center for Biotechnology Information (NCBI)
under the ascension number PRJNA1116256 or at the following URL:
https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1116256/.
Authors' contributions
DH designed the present study. SL performed the
sample and data detection. DH, SL and YZ analysed the data. SL
wrote the manuscript and participated in the literature collection
and evaluation. DH and SL confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The studies involving human participants were
reviewed and approved (approval no. IIT20220714A) by the
Institutional Review Board of the First Affiliated Hospital,
Zhejiang University School of Medicine (Hangzhou, China).
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of the present case report and any
accompanying images. The patient came from the First Affiliated
Hospital, Zhejiang University School of Medicine.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torres A, Chalmers JD, Dela Cruz CS,
Dominedò C, Kollef M, Martin-Loeches I, Niederman M and Wunderink
RG: Challenges in severe community-acquired pneumonia: A
point-of-view review. Intensive Care Med. 45:159–171.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Castillo NE, Rajasekaran A and Ali SK:
Legionnaires' disease: A review. Infect Dis Clin Pract.
24(1)2016.
|
|
3
|
Cunha BA, Nausheen S and Busch L: Severe Q
fever community-acquired pneumonia (CAP) mimicking Legionnaires'
disease: Clinical significance of cold agglutinins, anti-smooth
muscle antibodies and thrombocytosis. Heart Lung. 38:354–362.
2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Viasus D, Di Yacovo S, Garcia-Vidal C,
Verdaguer R, Manresa F, Dorca J, Gudiol F and Carratalà J:
Community-acquired legionella pneumophila pneumonia: A
single-center experience with 214 hospitalized sporadic cases over
15 years. Medicine (Baltimore). 92:51–60. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
García-Somoza MD, Fernández A, Prats E and
Verdaguer R: Community-acquired pneumonia caused by legionella
longbeachae serogroup 1 in an immunocompetent patient. Enferm
Infecc Microbiol Clín. 28:398–399. 2010.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
6
|
Snelling WJ, Sleator RD, Carrillo CD,
Lowery C, Moore JE, Pezacki JP and Dooley J: Current and Emerging
Microbiology Issues of Potable Water in Developed Countries. In:
Drinking Water: Contamination, Toxicity and Treatment. Molina SP
and Romero JD (eds). Nova Science Publishers, pp121-152, 2008.
|
|
7
|
Dagan A, Epstein D, Mahagneh A, Nashashibi
J, Geffen Y, Neuberger A and Miller A: Community-acquired versus
nosocomial legionella pneumonia: Factors associated with
legionella-related mortality. Eur J Clin Microbiol Infect Dis.
40:1419–1426. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pérez-Cobas AE, Ginevra C, Rusniok C,
Jarraud S and Buchrieser C: Persistent legionnaires' disease and
associated antibiotic treatment engender a highly disturbed
pulmonary microbiome enriched in opportunistic microorganisms.
mBio. 11:e00889–20. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pouderoux C, Ginevra C, Descours G, Ranc
AG, Beraud L, Boisset S, Magand N, Conrad A, Bergeron-Lafaurie A,
Jarraud S and Ader F: Slowly or nonresolving legionnaires' disease:
Case series and literature review. Clin Infect Dis. 70:1933–1940.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
David S, Rusniok C, Mentasti M,
Gomez-Valero L, Harris SR, Lechat P, Lees J, Ginevra C, Glaser P,
Ma L, et al: Multiple major disease-associated clones of legionella
pneumophila have emerged recently and independently. Genome Res.
26:1555–1564. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Han D, Yu F, Zhang D, Yang Q, Shen R,
Zheng S and Chen Y: Applicability of bronchoalveolar lavage fluid
and plasma metagenomic next-generation sequencing assays in the
diagnosis of pneumonia. Open Forum Infect Dis.
11(ofad631)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Han D, Yu F, Zhang D, Yang Q, Xie M, Yuan
L, Zheng J, Wang J, Zhou J, Xiao Y, et al: The real-world clinical
impact of plasma mNGS testing: An observational study. Microbiol
Spectr. 11(e0398322)2023.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
13
|
Fields BS, Benson RF and Besser RE:
Legionella and legionnaires' disease: 25 years of investigation.
Clin Microbiol Rev. 15:506–526. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
O'Brien SJ and Bhopal RS: Legionnaires'
disease: The infective dose paradox. Lancet. 342:5–6.
1993.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gu W, Miller S and Chiu CY: Clinical
metagenomic next-generation sequencing for pathogen detection. Annu
Rev Pathol. 14:319–338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
NNDSS Annual Report Writing Group.
Australia's notifiable disease status, 2015: Annual report of the
national notifiable diseases surveillance system. Commun Dis Intell
(2018). 43:2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Phin N, Parry-Ford F, Harrison T, Stagg
HR, Zhang N, Kumar K, Lortholary O, Zumla A and Abubakar I:
Epidemiology and clinical management of legionnaires' disease.
Lancet Infect Dis. 14:1011–1021. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Priest PC, Slow S, Chambers ST, Cameron
CM, Balm MN, Beale MW, Blackmore TK, Burns AD, Drinković D, Elvy
JA, et al: The burden of legionnaires' disease in New Zealand
(LegiNZ): A national surveillance study. Lancet Infect Dis.
19:770–777. 2019.PubMed/NCBI View Article : Google Scholar
|