Introduction
Psychotropic drugs are widely regarded as the
cornerstone of mental illness treatment (1,2).
Antidepressant therapy can typically occur for 2-3 years, whereas
maintenance therapy with antipsychotics may occur for up to 5
years; in certain cases, maintenance therapy may require lifelong
administration (3). However,
variations in pharmacokinetics and pharmacodynamics between
patients may lead to suboptimal responses or increased
susceptibility to adverse effects, even when standard therapeutic
doses are used (4,5). Phillips et al (6) reported that interindividual
differences in drug metabolism may contribute to nearly half of all
adverse drug reactions (7).
Although pharmacogenetic testing has demonstrated its potential for
increasing the efficacy and safety of psychotropic drugs by
identifying genetic polymorphisms linked to drug metabolism
(8-10),
its widespread application in mental health care is limited by high
costs. Therefore, there is need to develop affordable and effective
tools for evaluating metabolic capacities for antipsychotic
medications to guide clinical decision-making.
In addition to genetic factors, non-genetic factors
such as age, sex, weight, smoking status, comorbid disease and
liver and renal function have been shown to influence the
metabolism of antipsychotic drugs (11-14).
To the best of our knowledge, however, there are currently no
clinical prediction models based on these factors. Therefore, the
present study systematically collected patient data on
demographics, lifestyle habits, past medical history and
serological indicators to construct accurate clinical prediction
models for assessing metabolic capacities for psychotropic drugs.
Such models provide robust, evidence-based guidance for
personalized medication management. Multivariable logistic
regression analysis was initially performed on the total dataset to
identify potential risk factors associated with the metabolic
capacity for antipsychotics, antidepressants and anxiolytics. Based
on these significant factors, prediction models and nomograms were
developed for various psychotropic drugs. As a graphical predictive
tool, the nomogram provides a visual and intuitive approach to
forecast outcomes by integrating multiple influencing factors
(15). By transforming these
factors into intuitive predictive components, the nomogram
generates a total score that corresponds to the probability of a
specific outcome (16). This
personalized, user-friendly method provides an accessible way to
predict outcomes, enabling an intuitive prediction of ability to
metabolize psychotropic drugs. Additionally, the quality of the
prediction models was evaluated using multiple validation methods
to ensure their reliability and applicability in clinical
practice.
Materials and methods
Clinical specimens and study
design
Using the sample size formula for case-control study
design in PASS version 21.0.3 software (NCSS, LLC), a total sample
size of 112 participants (confidence level=0.95, power=0.8, assumed
odds ratio=4.0) was required to ensure the accuracy, reliability
and statistical power of the study results. As diabetes,
hypertension and liver and kidney function are associated with drug
metabolism, and these factors are important to consider when
building a model to predict drug metabolism, a broad range of
patients was included. Therefore, a total of 185 adult patients
(age, ≥18 years), who did not have cancer and were not critically
ill, with or without comorbidities such diabetes, hypertension and
liver and kidney diseases, and underwent pharmacogenomic testing at
The First Affiliated Hospital of Jilin University (Changchun,
China) between January 2022 and September 2023 were initially.
However, due to excessive missing clinical data, 31 patients were
excluded, resulting in 154 patients being available for analysis.
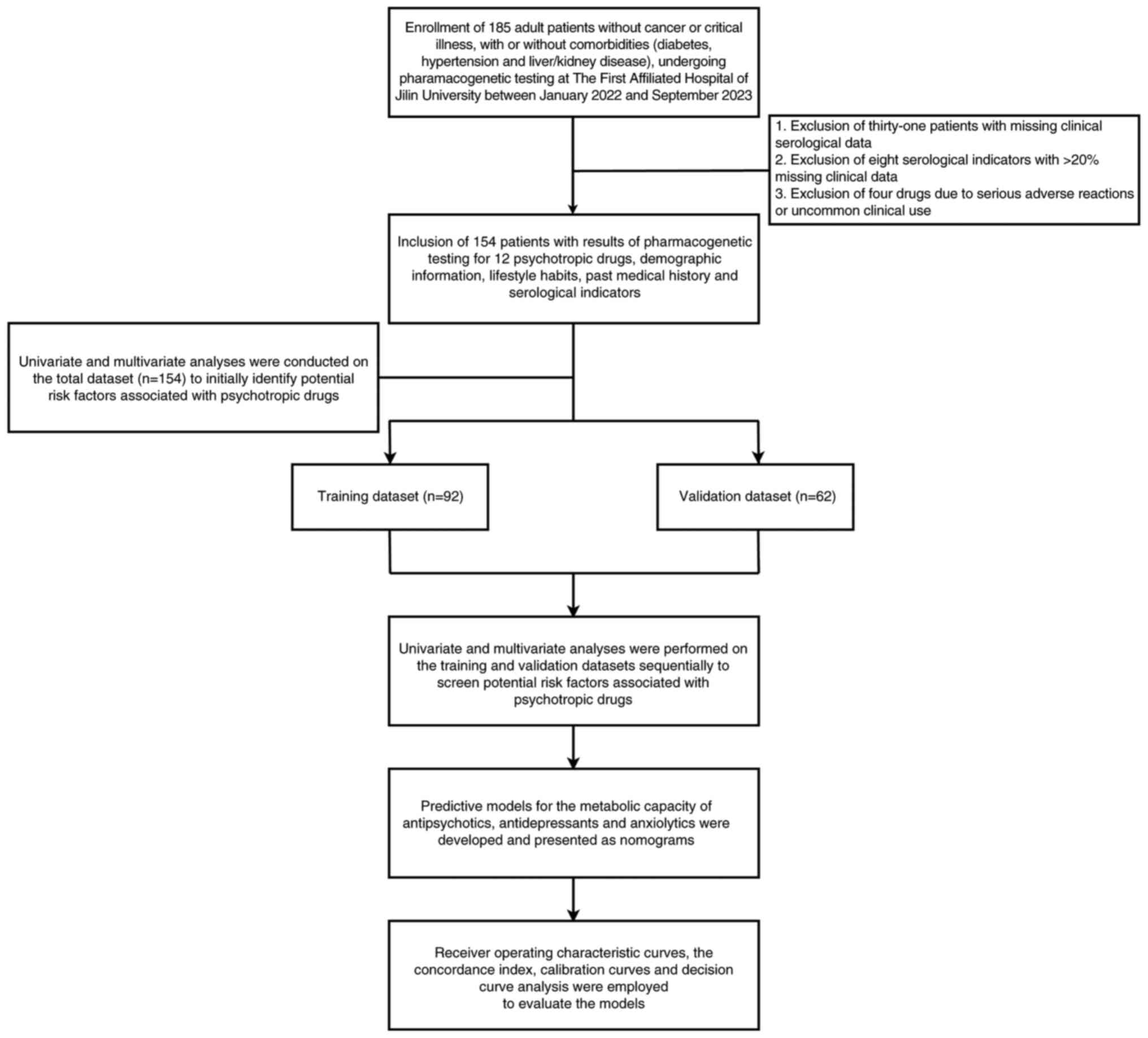
The dataset was initially used to screen potential risk factors and
was subsequently split into a training dataset and a validation
dataset at a 6:4 ratio. Variables that were identified as being
significant following multivariable adjustments were incorporated
into the final predictive models.
Clinical information, including pharmacogenetic
testing results for psychotropic drugs (four antipsychotics, ten
antidepressants and two anxiolytics), demographic information [age,
sex and body mass index (BMI)], lifestyle habits (smoking and
alcohol consumption status), past medical history (hypertension and
diabetes) and serological indicators [liver and renal function,
fasting blood glucose (FBG), blood lipids and hypersensitive
C-reactive protein (hs-CRP)], was collected from the hospital
records of all patients. The present study was approved by the
Ethics Committee of the First Affiliated Hospital of Jilin
University (approval no. AF-IRB-032-06), and due to its
retrospective nature, the requirement for informed consent was
waived. Psychotropic drugs associated with serious adverse
reactions, as well as those that are not commonly used in clinical
practice, including one antipsychotic (clozapine) and three
antidepressants (amitriptyline, doxepin and desipramine), were
excluded from the study. Furthermore, serological indicators that
are infrequently measured by clinical laboratories, including the
glomerular filtration rate, α-L-fucosidase, collagen type IV,
glycocholic acid, adenosine deaminase, monoamine oxidase,
5'-nucleotidase and cystatin C, were excluded. Missing values in
the remaining dataset were addressed via multiple imputation; mean
values obtained following five rounds of imputation were used for
analysis. There were 125 male and 29 female participants, with a
median age of 61 years (range, 20-91 years). After rigorous
screening (excluding patients with excessive missing clinical
information, drugs with uncommon use or severe adverse reactions,
as well as serological indicators that are infrequently measured)
and processing (imputing missing values), the dataset contained 154
patients and 31 clinical indicators [age, sex, BMI, smoking and
alcohol consumption status, hypertension, diabetes, hs-CRP,
homocysteine, FBG, triglyceride, total cholesterol, high-density
lipoprotein cholesterol (HDL-c), low-density lipoprotein
cholesterol, aspartate aminotransferase (AST), alkaline phosphatase
(ALP), alanine aminotransferase (ALT), γ-glutamyl transpeptidase
(GGT), cholinesterase (CHE), prealbumin, total protein, albumin,
globulin (GLOB), albumin to GLOB ratio (AGR), total (TBIL),
conjugated and unconjugated bilirubin, total bile acid (TBA), urea,
uric acid (UA) and creatinine]. These indicators were analysed for
potential associations with the metabolic capacity of 12
psychotropic drugs (three antipsychotics: Haloperidol, olanzapine
and risperidone; seven antidepressants: Citalopram, escitalopram,
sertraline, bupropion, paroxetine, mirtazapine and venlafaxine and
two anxiolytics: Oxazepam and lorazepam).
To enhance clinical applicability, continuous
variables (age, BMI and serological indicators) were converted into
categorical variables based on established criteria for statistical
analysis (17-21).
As age follows a normal distribution, the mean age (60.57) was used
as the cut-off point to divide the patients into younger (age ≤60
years) and older (age >60 years) groups. BMI categories were
classified according to Working Group on Obesity in China criteria
as follows: Underweight (BMI <18.5), normal weight (BMI ≥18.5
and <25), overweight (BMI ≥25) and obese (BMI ≥28
kg/m2) groups (17).
Similarly, serological indicators were categorized as normal,
decreased or elevated based on the standardized clinical ranges
reported by First Hospital of Jilin University (18-21).
Screening potential indicators related
to the abnormal metabolism of psychotropic drugs
A case-control analysis study design was utilized to
individually analyse psychotropic drugs. For each analysis,
individuals with normal metabolic capacities were classified as the
control group, whereas those with abnormal metabolism capacity were
designated the case group. Abnormal metabolism of psychotropic
drugs refers to genetic variations or polymorphisms that influence
drug metabolism at the genetic level. These variations impact the
activity, expression or function of drug metabolic enzymes, thereby
leading to abnormal drug metabolism (22).
To identify clinical indicators associated with drug
metabolism, uni- and multivariate analyses on the total dataset
were performed. Univariate analyses were conducted using the
unpaired two-independent sample t-tests for continuous variables
and χ2 tests for categorical variables. Multivariate
analyses were performed using three multivariable logistic
regression models to investigate the association between clinical
indicators and psychotropic drug metabolism. Clinical indicators
that were not significant according to χ2 test or
multivariable logistic regression were excluded from subsequent
analyses.
In the training dataset, significant clinical
indicators were rescreened to identify potential risk factors. The
associations between risk factors and differences in drug
metabolism were validated using the validation dataset. A schematic
overview of the study design is shown in Fig. 1.
Statistical analysis
Data are presented as the mean ± SD. In the
preliminary analysis of the data, the unpaired two-independent
sample t-tests were used to compare continuous variables, whereas
the categorical variables were assessed using χ2 or
Fisher's exact test, as appropriate. Potential risk factors
associated with abnormal psychotropic metabolism were further
evaluated via three multivariable binary logistic regression
models, with adjustments for two covariates (age and sex). Model 1
used binary logic regression to evaluate the association between
each clinical indicator and the metabolic capacity of psychotropic
drugs. In Model 2, variables that exhibited statistical
significance were selected for multivariable logistic regression
analysis. In Model 3, all clinical indicators were included in a
multivariable binary logistic regression analysis. P<0.05 was
considered to indicate a statistically significant difference. All
analyses were conducted in R (version 4.1.2, http://www.R-project.org/) and SPSS (version 26, IBM
Corp.).
Establishment and evaluation of
predictive models
Variables that were identified as statistically
significant were incorporated into the final multivariable model.
To display the results of the logistic regression, the coefficients
that were obtained from the multivariable analysis were used as
weights to develop a nomogram, which facilitates the practical
application of the model in evaluating the probability of abnormal
drug metabolism. The performance of the predictive model was
evaluated via receiver operating characteristic curves, the
concordance index, calibration curves and decision curve analysis
(DCA). The R packages used were ‘rms’ (version 6.7-0, http://cran.r-project.org/web/packages/rms), ‘pROC’
(version 1.18.4, http://cran.r-project.org/web/packages/pROC), and
‘rmda’ (version 1.6, http://cran.r-project.org/web/packages/rmda).
Results
Demographic and clinical
characteristics of the patients
A total of 154 patients with clinical information
and pharmacogenetic testing results were included. The
sociodemographic characteristics of the study sample are shown in
Tables SI, SII and SIII. Metabolic capacities for various
psychotropic drugs were estimated based on pharmacogenetic testing
(Table SIV). The majority of
patients exhibited normal metabolic capacities for haloperidol
(n=97; 63.0%), olanzapine (n=138; 89.6%), risperidone (n=148;
96.1%), citalopram/escitalopram (n=137; 89.0%), sertraline (n=141;
91.6%), and mirtazapine/venlafaxine (n=90; 58.4%). However, a
notable proportion of the patients displayed abnormal metabolism
for bupropion (n=107; 69.5%), paroxetine (n=72; 46.8%), and
oxazepam/lorazepam (n=120; 77.9%).
Preliminary identification of clinical
indicators associated with the ability to metabolize psychotropic
drugs via univariate analysis
Univariate analyses revealed clinical indicators
that were significantly associated with the ability to metabolize
psychotropic drugs in the total dataset (Tables SI, SII and SIII). These indicators included BMI,
hypertension, diabetes, hs-CRP, FBG, triglyceride, AST, ALT, γ-GGT,
CHE, prealbumin, unconjugated bilirubin and creatinine.
Screening of potential risk factors
associated with the ability to metabolize psychotropic drugs via
multivariate analysis
To investigate the independent risk factors
associated with abnormal drug metabolism, multivariate analyses
were conducted on the total dataset (Fig. 2A; Tables SV, SVI, SVII, SVIII, SIX, SX, SXI, SXII and SXIII). The multivariate analyses
revealed associations between clinical indicators and abnormal
metabolism of psychotropic drugs. For antipsychotics, metabolic
abnormality was significantly associated with BMI and ALP, γ-GGT,
CHE and UA levels for haloperidol, TBA levels for olanzapine and
GLOB and TBIL levels for risperidone. For antidepressants,
metabolic abnormality was significantly associated with BMI and ALP
levels for citalopram/escitalopram, BMI and FBG levels for
sertraline, urea and UA levels for bupropion, hypertension and AST
and CHE levels for paroxetine and hs-CRP, FBG, HDL-c, AST, ALT,
ALP, CHE and UA levels, as well as AGR and hypertension, for
mirtazapine/venlafaxine. For anxiolytics, abnormal metabolism of
oxazepam/lorazepam was associated with hypertension. These findings
were based on analyses conducted for individual drugs rather than
drug combinations.
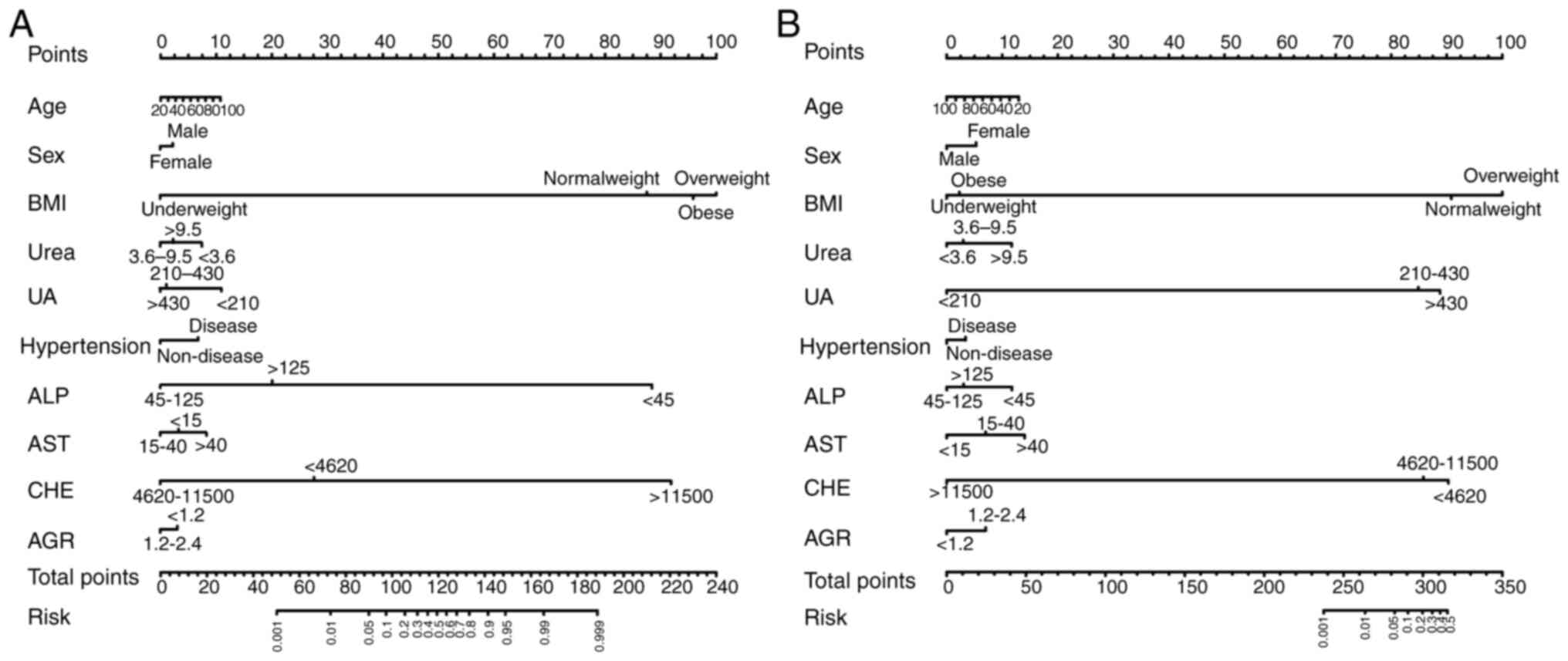
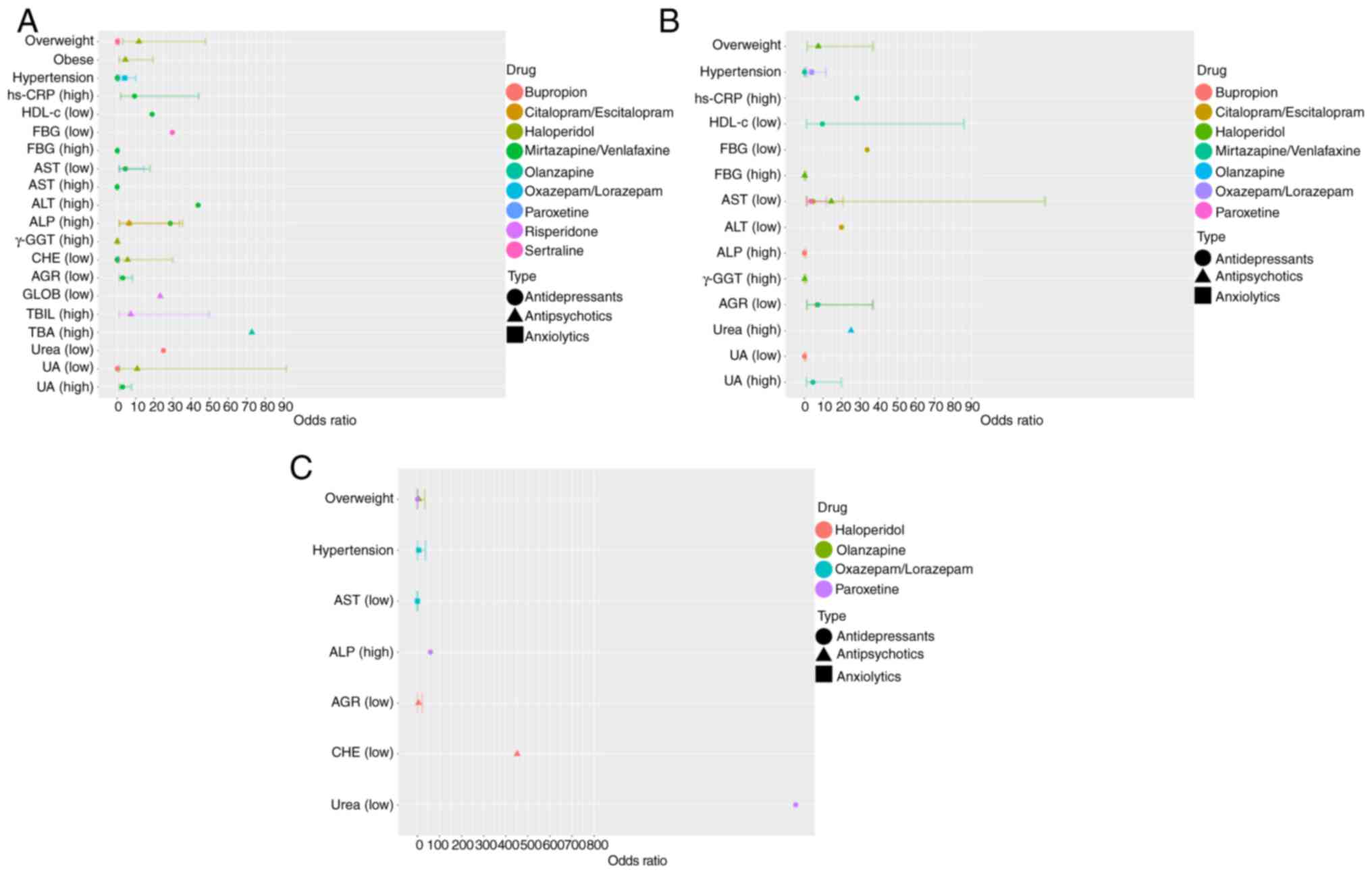 | Figure 2Multivariate logistic regression
analysis was used to analyse association between clinical
information, serological indicators and the metabolic capacity of
psychotropic drugs. Multivariate logistic regression analysis
results for (A) total, (B) training and (C) validation dataset.
hs-CRP, hypersensitive C-reactive protein; HDL-c, high-density
lipoprotein cholesterol; FBG, fasting blood glucose; AST, aspartate
aminotransferase; ALT, alanine aminotransferase; ALP, alkaline
phosphatase; GGT, glutamyl transpeptidase; CHE, cholinesterase;
AGR, albumin to globulin ratio; GLOB, globulin; TBIL, total
bilirubin; TBA, total bile acid; UA, uric acid. |
To verify the predictive ability, potential risk
factors were screened in the training dataset and association
between these factors and drug metabolism were assessed in the
validation dataset (Fig. 2B and
C; Tables SXIV, SXV, SXVI, SXVII, SXVIII, SXIX, SXX, SXXI, SXXII, SXXIII, SXXIV, SXXV, SXXVI, SXXVII, SXXVIII, SXXIX, SXXX and SXXXI). AGR, CHE, urea and UA emerged as
candidate risk factors via χ2 test or binary logistic
regression in the total and training dataset, whereas BMI,
hypertension, AST and ALP were significant indicators via both
χ2 test and binary logistic regression in the validation
dataset.
The multivariate analyses revealed several factors
related to the abnormal metabolism of psychotropic drugs.
Overweight individuals had significantly increased risk of abnormal
metabolism of haloperidol (OR, 7.493, 95% CI: 1.516-37.027), with
an increased risk also observed when CHE levels were decreased (OR,
5.668, 95% CI: 1.068-30.085). Similarly, overweight status was also
associated with abnormal metabolism of olanzapine (OR, 5.863, 95%
CI: 1.026-33.501). Moreover, a decrease in the AST levels was
identified as being a significant risk factor for abnormal
metabolism of paroxetine (OR, 4.404, 95% CI: 1.335-14.533).
Hypertension was a risk factor for abnormal
metabolism of oxazepam/lorazepam (OR, 3.878, 95% CI, 1.290-11.661)
but paradoxically acted as a protective factor for abnormal
metabolism of mirtazapine/venlafaxine (OR, 0.052, 95% CI:
0.01-0.278). Additionally, decreased levels of HDL-c (OR, 18.903,
95% CI: 1.301-274.721) and AGR (OR, 3.071, 95% CI: 1.139-8.281), as
well as increased levels of UA (OR, 4.597, 95% CI: 1.059-19.962),
were risk factors for abnormal metabolism of
mirtazapine/venlafaxine. Furthermore, multivariable logistic
regression analyses revealed significant associations between ALP
and urea and abnormal metabolism of both antipsychotics and
antidepressants.
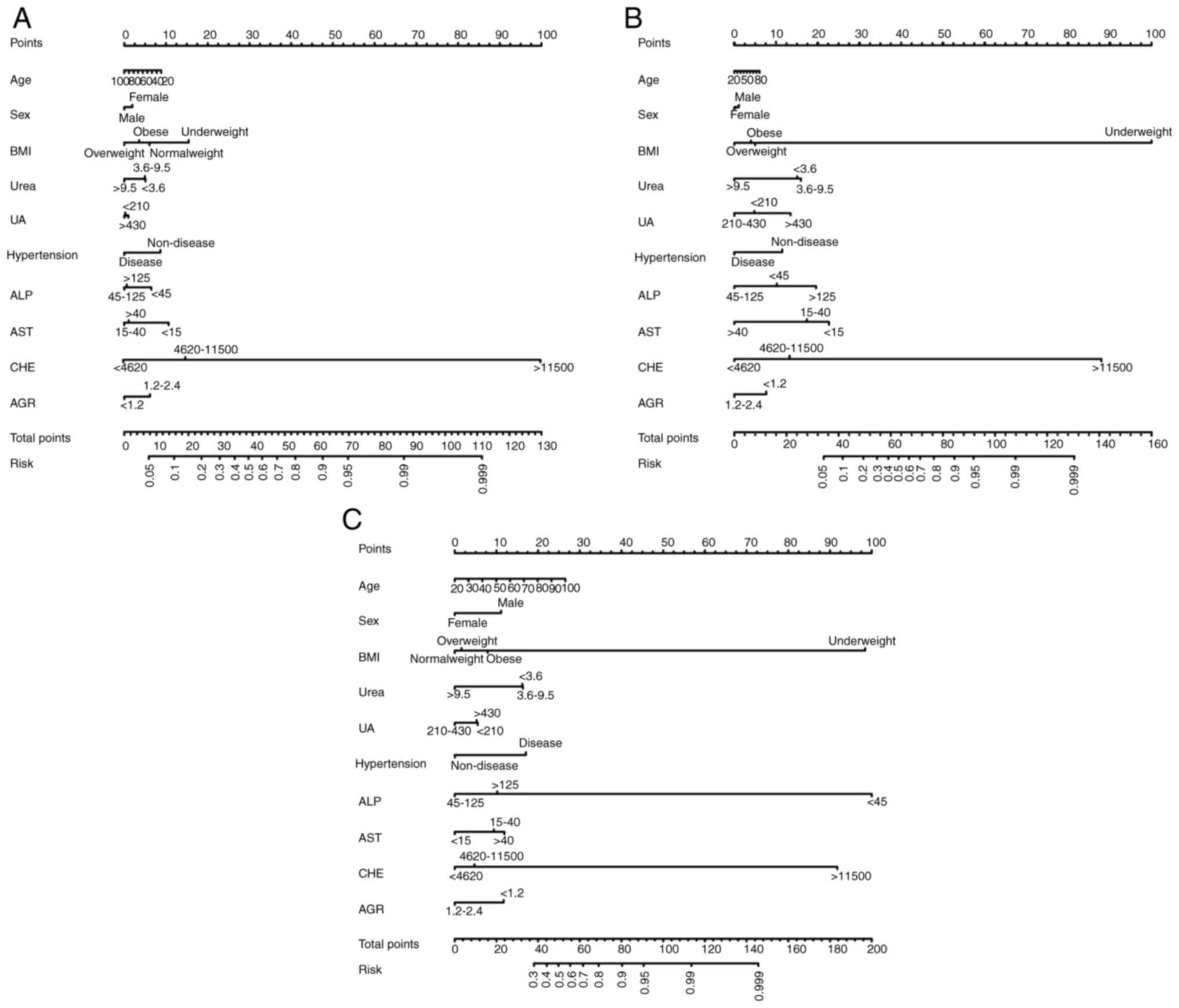
Feature selection and development of
individualized prediction models
Significant differences were observed in BMI,
hypertension and ALP, AST, CHE, AGR, urea and UA levels between
individuals with abnormal and normal metabolism. Using these
significant factors, along with age and sex, prediction models were
developed to assess the likelihood of abnormal metabolism for five
psychotropic drugs (Figs. 3 and
4).
The nomogram of haloperidol highlighted BMI as the
most influential factor in determining metabolic capacity, followed
by CHE and ALP. By contrast, sex and AGR exhibited minimal effects
on haloperidol metabolism. Similarly, the nomogram of olanzapine
indicated that BMI plays a pivotal role in determining metabolic
capacity, with CHE and UA also notably contributing, whereas
hypertension and sex exerted a limited influence. In the nomograms
of paroxetine, mirtazapine/venlafaxine and oxazepam/lorazepam,
various factors were also found to influence drug metabolism.
Specifically, in the case of paroxetine, CHE exhibited the most
prominent impact, followed by BMI and AST, with sex and UA
demonstrating limited effects. Similarly, the nomogram for
mirtazapine/venlafaxine highlighted BMI as the primary influencing
factor, followed by CHE and AST, whereas sex and age had minimal
impacts. Finally, the nomogram for oxazepam/lorazepam indicated
that ALP exhibited the greatest impact on metabolic capacity,
followed by BMI and CHE, whereas UA and sex had limited
impacts.
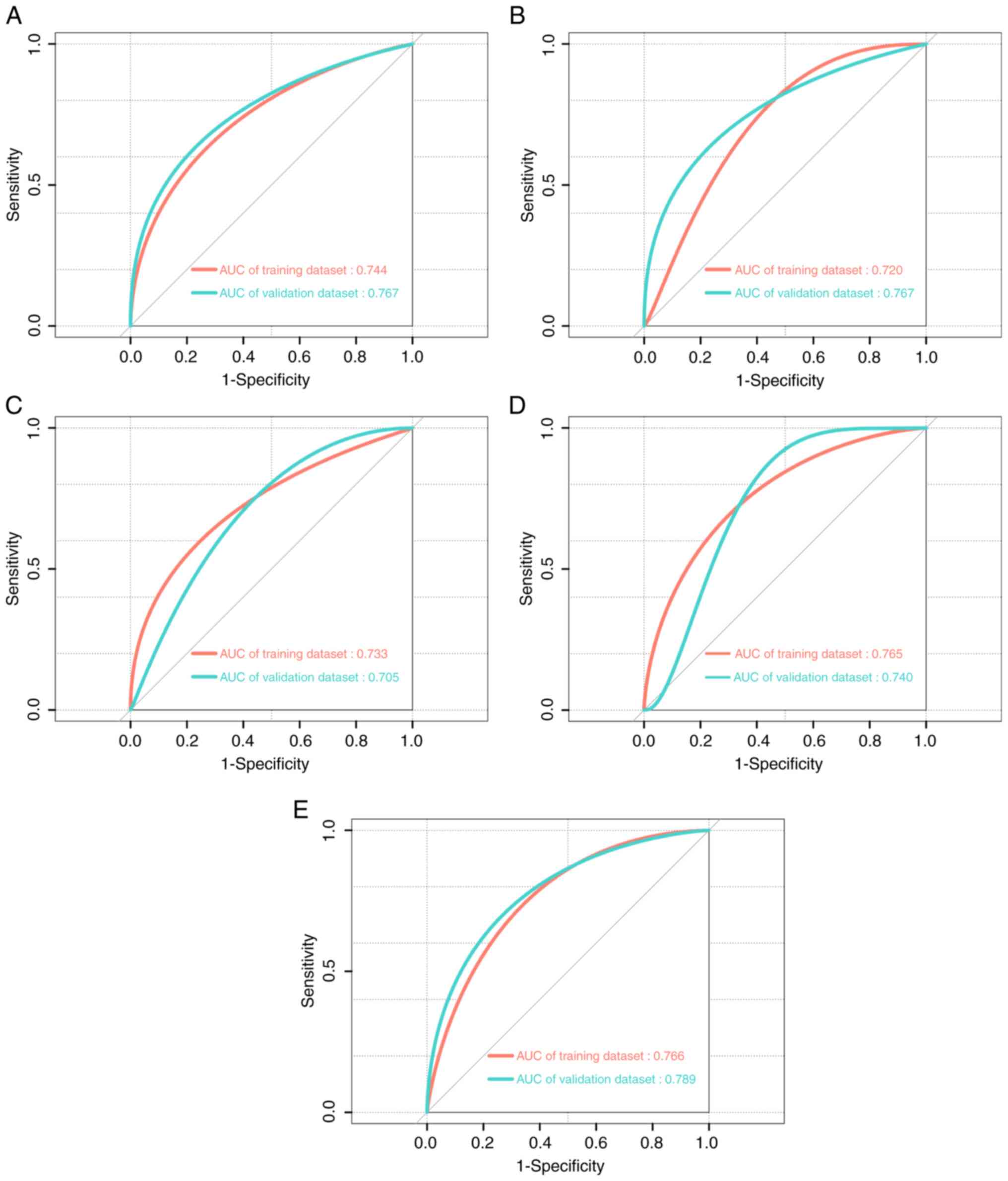
Performance evaluation of
individualized prediction models
The developed models incorporated age, sex, BMI,
hypertension, ALP, AST, CHE, AGR, urea and UA. For the 154
patients, the C-indices of the nomograms for haloperidol,
olanzapine, paroxetine, mirtazapine/venlafaxine and
oxazepam/lorazepam were 0.748, 0.733, 0.678, 0.734 and 0.751,
respectively. Additionally, in the training dataset, the area under
the receiver operating characteristic curve (AUC) values were
0.744, 0.720, 0.733, 0.765 and 0.766, respectively. In the
validation dataset, AUC values for these drugs were 0.767, 0.767,
0.705, 0.740, and 0.789, respectively (Fig. 5). These results indicated that the
models exhibited moderate predictive accuracy in predicting the
abnormal metabolic capacity of psychotropic drugs. To assess model
performance, calibration curves were used to evaluate the accuracy
of the nomograms (Figs. S1 and
S2). The predictive performance of
the nomograms in both the training and validation datasets was
moderately consistent with the actual results. In addition, DCA has
also been widely used to evaluate the clinical utility of nomograms
(23,24). As shown by the DCA, the nomograms
demonstrated a notable positive net benefit for predicting the risk
of abnormal metabolism capacity of psychotropic drugs, thus
indicating their potential for clinical application (Fig. S3).
Discussion
The present study developed and validated innovative
predictive models for assessing the metabolic capacity of
psychotropic drugs based on clinical characteristics and
serological indicators. These models provide a practical
alternative to pharmacogenetic testing by offering greater
accessibility, cost-effectiveness and time efficiency. Unlike
genetic testing, which requires advanced technology and equipment,
the present models rely on easily available clinical information,
thus making them more suitable for routine clinical use. Each of
the predictive models incorporated ten key variables that notably
influence the metabolism of psychotropic drugs, including age, sex,
BMI, hypertension, ALP, AST, CHE, AGR, urea and UA.
Elderly individuals are particularly susceptible to
the side effects of psychotropic drugs due to physiological,
pharmacokinetic and pharmacodynamic changes associated with age
(25). As age increases, DNA
hypermethylation typically occurs in the promoter region. This
epigenetic change results in decreased expression levels of genes
regulated by DNA methylation, including those involved in drug
metabolism and distribution, such as cytochrome P450 (CYP) enzymes;
these changes impair the metabolism of psychotropic drugs (26,27).
Previous studies have demonstrated that age affects the clearance
of escitalopram, whereas BMI affects the volume of distribution of
escitalopram (28-30).
The CYP450 enzyme family serves a central role in the metabolism of
psychotropic drugs. Several studies have shown that genetic
polymorphisms in CYP2C19, CYP2D6 and other genes are associated
with antipsychotic medication concentrations in the blood (31-34).
Although there is no significant sex-specific difference in CYP2C19
activity, it has been reported that female patients tend to exhibit
increased CYP2D6 activity (35,36).
Furthermore, patients with depression and certain types of anxiety
disorder (such as generalized anxiety disorder) have a notably
increased likelihood of concurrently developing hypertension
(37). Additionally, variations in
liver and renal function affect the pharmacokinetics of individuals
(38,39). Depression is associated with an
increase in oxidative stress and a decrease in antioxidant defences
(40). Chaudhari et al
(41) reported a negative
association between serum UA levels and the intensity of
depression, which may be attributed to the potent antioxidant
properties of UA.
The present study established effective nomograms
that display the impacts of risk factors on drug metabolism and
effectively evaluate metabolic capacities for psychotropic drugs.
These nomograms incorporate easily accessible clinical indicators,
thereby enhancing their practicality and clinical utility. These
nomograms predict the possibility of abnormal drug metabolism in
patients, thus providing clinical decision-making references for
the necessity of early screenings for psychiatric drug metabolism
and pharmacogenetic testing. For patients with a low predicted
likelihood of abnormal metabolism, pharmacogenetic testing can be
deferred or cancelled, thus conserving both time and resources.
Within-cohort validation demonstrated that the models possessed
reasonable discriminatory and calibration abilities.
However, the present study had limitations. First,
the retrospective study design and relatively small sample size may
introduce a potential selection bias. Second, the estimated
glomerular filtration rate, the main index for evaluating renal
function, was not included in the logistic regression analysis due
to the lack of available data in the retrospective study. Follow-up
studies incorporating more appropriate variables are needed to
refine the model and enhance its ability to assess abnormal drug
metabolism. Third, although the robustness of nomograms was
internally validated in the same population, external validation in
populations from other regions and countries is lacking.
Multicentre studies with larger, more diverse populations are
needed to validate and generalize the predictive models.
In conclusion, BMI, hypertension, ALP, AST, CHE,
AGR, urea and UA serve key roles in distinguishing between normal
and abnormal metabolism of antipsychotic drugs. Based on these
associations, convenient, cost-effective, non-invasive, radiation
exposure risk-free and simple prediction models were established.
The nomograms constructed based on these models exhibited
favourable diagnostic accuracy and calibration in distinguishing
between normal and abnormal drug metabolism. To extend the
applicability of these models and promote their use in diverse
populations, further validation with data from different
institutions is warranted.
Supplementary Material
Calibration curve analysis of the
nomograms in the training dataset. (A) Haloperidol. (B) Olanzapine.
(C) Paroxetine. (D) Mirtazapine/venlafaxine. (E)
Oxazepam/lorazepam.
Calibration curve analysis of the
nomograms in the validation dataset. (A) Haloperidol. (B)
Olanzapine. (C) Paroxetine. (D) Mirtazapine/venlafaxine. (E)
Oxazepam/lorazepam.
Decision curve analysis of the
nomograms in the training and validation datasets. (A) Haloperidol.
(B) Olanzapine. (C) Paroxetine. (D) Mirtazapine/venlafaxine. (E)
Oxazepam/lorazepam.
Risk factors that affect ability to
metabolize antipsychotics.
Risk factors that affect ability to
metabolize antidepressants.
Risk factors that affect ability to
metabolize anxiolytics (oxazepam/lorazepam).
Pharmacogenetic testing of
psychotropic drugs.
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the total dataset (adjusted by age and
sex).
Multiple logistic regression results
of abnormal metabolism of antidepressants associated with clinical
indicators based on the total dataset (adjusted by age and
sex).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the total dataset
(adjusted by age and sex).
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the total dataset (adjusted by significant
factor in the χ2 or Fisher’s exact tests).
Multiple logistic regression results
of abnormal metabolism of antidepressants associated with clinical
indicators based on the total dataset (adjusted by the significant
factor in χ2 tests).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the total dataset
(adjusted by the significant factor in χ2 tests).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
antipsychotics based on the total dataset (adjusted by all clinical
indicators).
Multiple logistic regression results
of clinical indexes associated with abnormal drug metabolism in
antidepressants based on the total dataset (adjusted by all
clinical indicators).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
anxiolytics (oxazepam/lorazepam) based on the total dataset
(adjusted by all clinical indicators).
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the training dataset (adjusted by age and
sex).
Multiple logistic regression of
abnormal metabolism of antidepressants associated with clinical
indicators based on the training dataset (adjusted by age and
sex).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the training dataset
(adjusted by age and sex)
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the training dataset (adjusted by the
significant factor in χ2 tests).
Multiple logistic regression results
of abnormal metabolism of antidepressants associated with clinical
indicators based on the training dataset (adjusted by the
significant factor in χ2 tests).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the training dataset
(adjusted by the significant factor in χ2 tests).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
antipsychotics based on the training dataset (adjusted by all
clinical indicators).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
antidepressants based on the training dataset (adjusted by all
clinical indicators).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
anxiolytics (oxazepam/lorazepam) based on the training dataset
(adjusted by all clinical indicators).
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the validation dataset (adjusted by age, sex
and BMI).
Multiple logistic regression results
of abnormal metabolism of antidepressants associated with clinical
indicators based on the validation dataset (adjusted by age, sex
and BMI).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the validation dataset
(adjusted by age, sex and BMI).
Multiple logistic regression results
of abnormal metabolism of antipsychotics associated with clinical
indicators based on the validation dataset (adjusted by the
significant factor in χ2 tests).
Multiple logistic regression results
of abnormal metabolism of antidepressants (mirtazapine/venlafaxine)
associated with clinical indicators based on the validation dataset
(adjusted by the significant factor in χ2 tests).
Multiple logistic regression results
of abnormal metabolism of anxiolytics (oxazepam/lorazepam)
associated with clinical indicators based on the validation dataset
(adjusted by the significant factor in χ2tests).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
antipsychotics based on the validation dataset (adjusted by all
clinical indicators).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
antidepressants based on the validation dataset (adjusted by all
clinical indicators).
Multiple logistic regression results
of clinical indexes related to abnormal drug metabolism in
anxiolytics (oxazepam/lorazepam) based on the validation dataset
(adjusted by all clinical indicators).
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by National Natural
Science Foundation of China (grant no. 22174137), Jilin Province
Science and Technology Agency (grant nos. YDZJ202501ZYTS066,
2023C013, JJKH20211210KJ, JJKH20211164KJ, JLSWSRCZX2020-009,
20200901025SF and 20200403084SF) and Beijing Medical Award
Foundation (grant no. YXJL-2021-1097-0645).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SZ, XH and YJ analyzed the data and wrote the
manuscript. PZ, JS and YJ conceived the study, supervised the
research and edited the manuscript. All authors have read and
approved the final manuscript. SZ and XH confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The Ethical Standard Research Committee of the First
Affiliated Hospital of Jilin University approved the present study
(approval no. AF-IRB-032-06). The requirement for informed consent
was waived due to the retrospective nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Goff DC: The pharmacologic treatment of
schizophrenia-2021. JAMA. 325:175–176. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Goff DC, Falkai P, Fleischhacker WW,
Girgis RR, Kahn RM, Uchida H, Zhao J and Lieberman JA: The
long-term effects of antipsychotic medication on clinical course in
schizophrenia. Am J Psychiatry. 174:840–849. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Middeldorp CM, Cath DC, Van Dyck R and
Boomsma DI: The co-morbidity of anxiety and depression in the
perspective of genetic epidemiology. A review of twin and family
studies. Psychol Med. 35:611–624. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Pardiñas AF, Owen MJ and Walters JTR:
Pharmacogenomics: A road ahead for precision medicine in
psychiatry. Neuron. 109:3914–3929. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Biso L, Aringhieri S, Carli M, Scarselli M
and Longoni B: Therapeutic drug monitoring in psychiatry: Enhancing
treatment precision and patient outcomes. Pharmaceuticals (Basel).
17(642)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Phillips KA, Veenstra DL, Oren E, Lee JK
and Sadee W: Potential role of pharmacogenomics in reducing adverse
drug reactions: A systematic review. JAMA. 286:2270–2279.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Solomon HV, Cates KW and Li KJ: Does
obtaining CYP2D6 and CYP2C19 pharmacogenetic testing predict
antidepressant response or adverse drug reactions? Psychiatry Res.
271:604–613. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chaplin M, Kirkham JJ, Dwan K, Sloan DJ,
Davies G and Jorgensen AL: STrengthening the reporting of
pharmacogenetic studies: Development of the STROPS guideline. PLoS
Med. 17(e1003344)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Verbelen M, Weale ME and Lewis CM:
Cost-effectiveness of pharmacogenetic-guided treatment: Are we
there yet? Pharmacogenomics J. 17:395–402. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Leitch TM, Killam SR, Brown KE, Katseanes
KC, George KM, Schwanke C, Loveland J, Elias AF, Haney K, Krebsbach
K, et al: Ensuring equity: Pharmacogenetic implementation in rural
and tribal communities. Front Pharmacol. 13(953142)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kloosterboer SM, Egberts KM, de Winter
BCM, van Gelder T, Gerlach M, Hillegers MHJ, Dieleman GC, Bahmany
S, Reichart CG, van Daalen E, et al: Pipamperone population
pharmacokinetics related to effectiveness and side effects in
children and adolescents. Clin Pharmacokinet. 59:1393–1405.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hassab Errasoul A and Alarabi MA: Factors
predicting serum clozapine levels in Middle Eastern patients: An
observational study. BMC Psychiatry. 22(269)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sun L, von Moltke L and Rowland Yeo K:
Application of physiologically based pharmacokinetic modeling to
predict the effect of renal impairment on the pharmacokinetics of
olanzapine and samidorphan given in combination. Clin
Pharmacokinet. 60:637–647. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li XL, Huang SQ, Xiao T, Wang XP, Kong W,
Liu SJ, Zhang Z, Yang Y, Huang SS, Ni XJ, et al: Pharmacokinetics
of immediate and sustained-release formulations of paroxetine:
Population pharmacokinetic approach to guide paroxetine
personalized therapy in chinese psychotic patients. Front
Pharmacol. 13(966622)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lin S, Zhu N, YihanZhang Du L and Zhang S:
Development and validation of a prediction model of
catheter-related thrombosis in patients with cancer undergoing
chemotherapy based on ultrasonography results and clinical
information. J Thromb Thrombolysis. 54:480–491. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jin H, Shen X, Song W, Liu Y, Qi L and
Zhang F: The development of nomograms to predict blastulation rate
following cycles of in vitro fertilization in patients with tubal
factor infertility, polycystic ovary syndrome, or endometriosis.
Front Endocrinol (Lausanne). 12(751373)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen C and Lu FC: Department of Disease
Control Ministry of Health, PR China. The guidelines for prevention
and control of overweight and obesity in Chinese adults. Biomed
Environ Sci. 17 (Suppl):S1–S36. 2004.PubMed/NCBI
|
|
18
|
National Health Commission of the People's
Republic of China: WS/T 404.1-2012 Reference Intervals for Common
Clinical Biochemical Tests-Part 1. China Standards Press, Beijing,
2012.
|
|
19
|
National Health Commission of the People's
Republic of China: WS/T 404.2-2012 Reference Intervals for Common
Clinical Biochemical Tests-Part 2. China Standards Press, Beijing,
2012.
|
|
20
|
National Health Commission of the People's
Republic of China: WS/T 404.4-2012 Reference Intervals for Common
Clinical Biochemical Tests-Part 4. China Standards Press, Beijing,
2012.
|
|
21
|
National Health Commission of the People's
Republic of China: WS/T 404.6-2012 Reference Intervals for Common
Clinical Biochemical Tests-Part 6. China Standards Press, Beijing,
2012.
|
|
22
|
Rajman I, Knapp L, Morgan T and
Masimirembwa C: African genetic diversity: Implications for
cytochrome P450-mediated drug metabolism and drug development.
EBioMedicine. 17:67–74. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Song C, Yu D, Wang Y, Wang Q, Guo Z, Huang
J, Li S and Hu W: Dual primary cancer patients with lung cancer as
a second primary malignancy: A population-based study. Front Oncol.
10(515606)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang S, Tian S, Li Y, Zhan N, Guo Y, Liu
Y, Xu J, Ma Y, Zhang S, Song S, et al: Development and validation
of a novel scoring system developed from a nomogram to identify
malignant pleural effusion. EBioMedicine. 58(102924)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lozupone M, Panza F, Stella E, La Montagna
M, Bisceglia P, Miscio G, Galizia I, Daniele A, di Mauro L, Bellomo
A, et al: Pharmacogenetics of neurological and psychiatric diseases
at older age: Has the time come? Expert Opin Drug Metab Toxicol.
13:259–277. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gomez A and Ingelman-Sundberg M:
Epigenetic and microRNA-dependent control of cytochrome P450
expression: A gap between DNA and protein. Pharmacogenomics.
10:1067–1076. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Seripa D, Panza F, Daragjati J, Paroni G
and Pilotto A: Measuring pharmacogenetics in special groups:
Geriatrics. Expert Opin Drug Metab Toxicol. 11:1073–1088.
2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bies RR, Feng Y, Lotrich FE, Kirshner MA,
Roose S, Kupfer DJ and Pollock BG: Utility of sparse concentration
sampling for citalopram in elderly clinical trial subjects. J Clin
Pharmacol. 44:1352–1359. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dhillon S, Scott LJ and Plosker GL:
Escitalopram: A review of its use in the management of anxiety
disorders. CNS Drugs. 20:763–790. 2006.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jin Y, Pollock BG, Frank E, Cassano GB,
Rucci P, Müller DJ, Kennedy JL, Forgione RN, Kirshner M, Kepple G,
et al: Effect of age, weight, and CYP2C19 genotype on escitalopram
exposure. J Clin Pharmacol. 50:62–72. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kirchheiner J, Nickchen K, Bauer M, Wong
ML, Licinio J, Roots I and Brockmöller J: Pharmacogenetics of
antidepressants and antipsychotics: The contribution of allelic
variations to the phenotype of drug response. Mol Psychiatry.
9:442–473. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Richards-Belle A, Austin-Zimmerman I, Wang
B, Zartaloudi E, Cotic M, Gracie C, Saadullah Khani N,
Wannasuphoprasit Y, Wronska M, Dawda Y, et al: Associations of
antidepressants and antipsychotics with lipid parameters: Do
CYP2C19/CYP2D6 genes play a role? A UK population-based study. J
Psychopharmacol. 37:396–407. 2023.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zanger UM and Schwab M: Cytochrome P450
enzymes in drug metabolism: Regulation of gene expression, enzyme
activities, and impact of genetic variation. Pharmacol Ther.
138:103–141. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Austin-Zimmerman I, Wronska M, Wang B,
Irizar H, Thygesen JH, Bhat A, Denaxas S, Fatemifar G, Finan C,
Harju-Seppänen J, et al: The influence of CYP2D6 and CYP2C19
genetic variation on diabetes mellitus risk in people taking
antidepressants and antipsychotics. Genes (Basel).
12(1758)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hägg S, Spigset O and Dahlqvist R:
Influence of gender and oral contraceptives on CYP2D6 and CYP2C19
activity in healthy volunteers. Br J Clin Pharmacol. 51:169–173.
2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Meibohm B, Beierle I and Derendorf H: How
important are gender differences in pharmacokinetics? Clin
Pharmacokinet. 41:329–342. 2002.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Maatouk I, Herzog W, Böhlen F, Quinzler R,
Löwe B, Saum KU, Brenner H and Wild B: Association of hypertension
with depression and generalized anxiety symptoms in a large
population-based sample of older adults. J Hypertens. 34:1711–1720.
2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Castberg I, Westin AA, Skogvoll E and
Spigset O: Effects of age and gender on the serum levels of
clozapine, olanzapine, risperidone, and quetiapine. Acta Psychiatr
Scand. 136:455–464. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shah RR: Drug development and use in the
elderly: Search for the right dose and dosing regimen (parts I and
II). Br J Clin Pharmacol. 58:452–469. 2004.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Black CN, Bot M, Scheffer PG, Cuijpers P
and Penninx BW: Is depression associated with increased oxidative
stress? A systematic review and meta-analysis.
Psychoneuroendocrinology. 51:164–175. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chaudhari K, Khanzode S, Khanzode S,
Dakhale G, Saoji A and Sarode S: Clinical correlation of alteration
of endogenous antioxidant-uric acid level in major depressive
disorder. Indian J Clin Biochem. 25:77–81. 2010.PubMed/NCBI View Article : Google Scholar
|