Introduction
Clinical and epidemiological studies suggest that
chronic inflammation predisposes individuals to different types of
cancer, and inflammatory molecules promote the proliferation of
malignant cells (1,2). The connection between inflammation
and cancer is mediated by several mechanisms, including genetic and
epigenetic alterations, that generate an inflammatory
microenvironment that further reinforces the development of cancer
(3). Moreover, functional
polymorphisms of inflammatory cytokine genes are associated with
cancer susceptibility (4–6).
Interleukin-6 (IL6) is a pleiotropic inflammatory
cytokine that is important for immune responses, cell survival,
proliferation and apoptosis (7).
Elevated expression of IL6 and its major effector, signal
transducer and activator of transcription-3 (STAT3), have been
implicated in different stages of tumor development, including
initiation, promotion, malignant conversion, invasion and
metastasis (8–12). The best characterized genetic
variants of IL6 is a G-to-C substitution at position -174,
upstream of the transcription start site, which has been reported
to influence IL6 levels in vitro and in vivo
(13,14). Elucidation of an association, if
any, between this polymorphism and cancer risk would support the
hypothesis that genetic variants in IL6, resulting in
aberrant IL6 expression, play a role in cancer development.
Individual studies and previously published
meta-analyses regarding the association of IL6 -174G/C with
cancer susceptibility (15,16)
enrolled too few subjects to provide conclusive evidence for or
against an association of this polymorphism with cancer risk. The
aim of this study was to assess the association of IL6
-174G/C polymorphism with cancer risk by conducting a comprehensive
meta-analysis of all eligible case-control studies.
Materials and methods
This meta-analysis was performed according to the
guidelines of the preferred reporting items for systematic reviews
and meta-analyses (PRISMA statement) (17) and the reporting meta-analysis of
observational studies in epidemiology (MOOSE) (18).
Data sources and study selection
To identify all studies on the association between
the IL6 -174G/C polymorphism and cancer risk, we conducted a
systematic search of the literature published before April 2011
using the MEDLINE database and the HuGE Published Literature
database (HuGE Pub Lit) (19) with
no restrictions. For MEDLINE, keywords ‘IL-6’ OR ‘IL 6’ OR ‘IL6’ OR
‘interleukin-6’ OR ‘interleukin 6’ AND ‘polymorphism’ AND ‘cancer’
were used; For HuGE Pub Lit, keywords ‘IL6’ AND ‘cancer’ were used
for searching eligible studies. In addition, a manual review of
references from primary or review articles was screened to trace
additional relevant studies. Studies were included if they had a
case-control design and the available frequency of three genotypes
regarding the IL6 -174G/C polymorphism. Of the studies with
overlapping data, we selected the ones with the largest number of
subjects.
Data extraction
Three investigators independently extracted data and
reached a consensus on all items. The following data were extracted
from each study: the first author's last name, publication year,
ethnicity of the subjects, cancer type, study design (retrospective
case-control or prospective cohort study), and numbers of genotyped
cases and controls with GG, GC or CC genotypes. Ethnic group was
defined as African, Caucasian or ‘mixed’, including more than one
ethnic category. Studies investigating more than one type of cancer
with overlapping or same controls were regarded as individual data
sets only in subgroup analyses by cancer type.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) analysis for the
frequencies of GG, GC and CC genotypes among controls in each study
was assessed using Pearson's Chi-square test. The strength of the
association between IL6 -174G/C polymorphism and cancer risk
was measured by odds ratio (OR) with its 95% confidence interval
(CI). The pooled ORs for IL6 -174G/C genotypes CC, GC and C
allele carriers (CC or GC) against GG genotype were calculated,
respectively. The significance of the pooled OR was determined by
the Z-test and P<0.05 was considered statistically significant.
Subgroup analysis was performed using stratification by study
character, cancer type, ethnicity and study design, respectively.
If a cancer type contained less than three independent individual
studies, it was categorized into the ‘other types’ group.
Testing for heterogeneity among studies was
performed by a Chi-square-based Q-test (20). Since Q-test is poor at detecting
true heterogeneity, heterogeneity was considered significant for
P<0.10 rather than P<0.05 (21). Additionally, the magnitude of the
between-study heterogeneity was also assessed by I2,
which can be calculated from the basic results of a typical
meta-analysis as I2 = 100% x (Q-df)/Q, ranges form 0 to
100%, and is typically considered low for I2<25%,
modest for 25–50% and large for >50% (22). Meta-regression was carried out to
investigate whether statistical heterogeneity between the results
of the multiple studies was related to one or more characteristics
of the studies (23). To identify
the studies that led to significant heterogeneity, sensitivity
analysis for between-study heterogeneity was implemented by the
sequential algorithm proposed by Patsopoulos et al (24). Whenever the P-value of the Q-test
was >0.10, the summarized OR estimate of each study was
calculated by the fixed-effects model (Mantel-Haenszel method)
(25). Otherwise, the
random-effects model (DerSimonian and Laird method) was used
(26). Funnel plots were used to
examine whether the results of a meta-analysis may have been
affected by publication bias (27). A modified version of Egger's test
proposed by Harbord, Egger and Sterne was implemented to test
funnel plot asymmetry (28). All
statistical analyses were performed using Stata statistical
software (Stata/SE version 10.1 for Windows; Stata Corp, College
Station, TX, USA).
Results
Characteristics of the included
studies
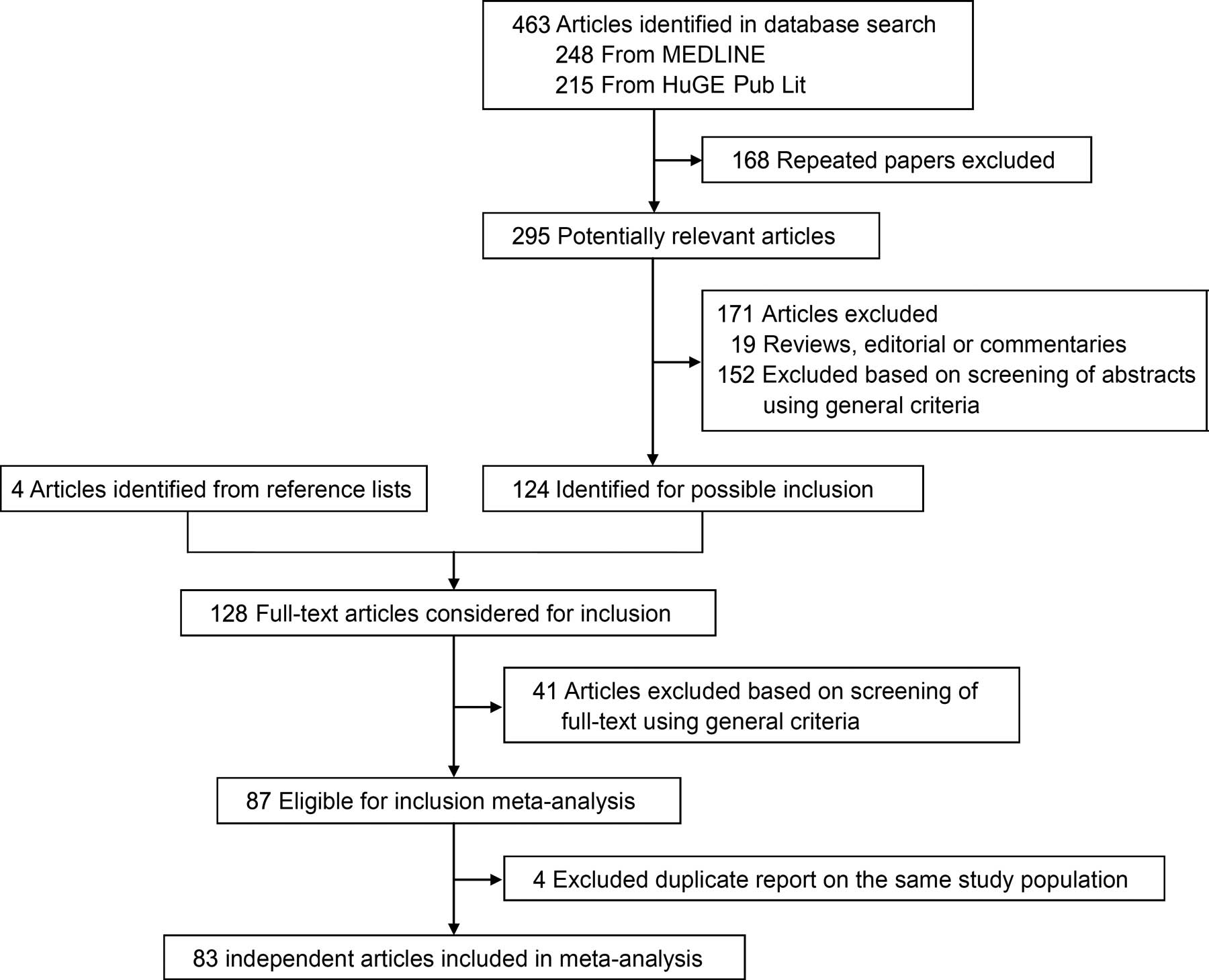
The detailed steps of our literature search are
described in Fig. 1. Eighty-three
independent articles that met the inclusion criteria were included
in the final analysis. Of these articles, one study provided data
on breast and prostate cancer using independent controls (29), therefore each group in this article
was treated as an independent study in our meta-analysis. The
characteristics of the included studies are summarized in Table I. Overall, the present
meta-analysis is based on a total of 105,482 participants,
including 44,735 cancer patients and 60,747 controls. The studies
were published between April 2000 and March 2011. Seventeen studies
were conducted with a prospective cohort design, and 67 were
conducted with a retrospective case-control design. Approximately
two-thirds of cases and controls (29,019 cases and 42,120 controls)
were from 73 studies involving Caucasian populations, a fraction of
the data (1,138 cases and 1,299 controls) from seven studies
involving African populations, and nearly one-third of the data
(14,578 cases and 17,328 controls) from four studies involving
‘mixed’ populations. As shown in Table
I, there were two studies from Dossus et al involving
Caucasians, African-Americans, Asians, Latinos and native
Hawaiians; one study from Ognjanovic et al involving
Caucasians, Asians and Hawaiians; and one study from Bushley et
al involving Caucasians, Asians and other populations. In the
controls, the frequency of the rare C allele among controls varied
considerably between Caucasians and Africans (0.417±0.052 and
0.207±0.097, respectively; P<0.001). A significant deviation
from HWE was noted in two studies in Africans, nine in Caucasians
and four in mixed populations (Table
I).
 | Table I.Characteristics of all studies
included in the meta-analysis. |
Table I.
Characteristics of all studies
included in the meta-analysis.
| Author | Year | Ethnicity | Cancer type | Study
designa | Cases
| Controls
| P-valueb |
|---|
| | | | | GG | GC | CC | GG | GC | CC | |
|---|
| Snoussi | 2005 | African | Breast cancer | Retrospective | 199 | 98 | 8 | 150 | 46 | 4 | 0.830 |
| Vishnoi | 2007 | African | Gallbladder
cancer | Retrospective | 97 | 25 | 2 | 153 | 44 | 3 | 0.936 |
| Ahirwar | 2008 | African | Bladder cancer | Retrospective | 86 | 24 | 26 | 130 | 56 | 14 | 0.027 |
| Kesarwani | 2008 | African | Prostate
cancer | Retrospective | 102 | 84 | 14 | 103 | 87 | 10 | 0.120 |
| Upadhyay | 2008 | African | Esophageal
cancer | Retrospective | 135 | 28 | 5 | 131 | 62 | 8 | 0.845 |
| Badr El-Din | 2009 | African | Brain tumor | Retrospective | 6 | 27 | 12 | 5 | 87 | 6 | 0.000 |
| Gangwar | 2009 | African | Cervical
cancer | Retrospective | 107 | 36 | 17 | 142 | 51 | 7 | 0.372 |
| Foster | 2000 | Caucasian | Kaposi sarcoma | Retrospective | 61 | 44 | 10 | 44 | 55 | 27 | 0.214 |
| Hulkkonen | 2000 | Caucasian | Leukemia | Retrospective | 8 | 13 | 14 | 81 | 201 | 118 | 0.785 |
| Zheng | 2000 | Caucasian | Multiple
myeloma | Retrospective | 22 | 36 | 15 | 33 | 69 | 26 | 0.357 |
|
Martinez-Escribano | 2002 | Caucasian | Melanoma | Retrospective | 14 | 26 | 2 | 20 | 26 | 2 | 0.071 |
| El-Omar | 2003 | Caucasian | Esophageal and
gastric cancer | Retrospective | 88 | 91 | 34 | 83 | 98 | 28 | 0.913 |
| Howell | 2003 | Caucasian | Melanoma | Retrospective | 48 | 79 | 34 | 79 | 101 | 44 | 0.258 |
| Hwang | 2003 | Caucasian | Gastric cancer | Retrospective | 19 | 9 | 2 | 22 | 8 | 0 | 0.399 |
| Landi | 2003 | Caucasian | Colorectal
cancer | Retrospective | 133 | 180 | 48 | 145 | 133 | 33 | 0.761 |
| Yakupova | 2003 | Caucasian | Multiple
myeloma | Retrospective | 23 | 33 | 13 | 37 | 53 | 12 | 0.286 |
| Campa | 2004 | Caucasian | Lung cancer | Retrospective | 64 | 111 | 68 | 55 | 105 | 47 | 0.818 |
| Cozen | 2004 | Caucasian | Lymphoma | Retrospective | 41 | 37 | 8 | 25 | 39 | 14 | 0.858 |
| Gazouli | 2004 | Caucasian | Kaposi sarcoma | Retrospective | 10 | 4 | 1 | 11 | 22 | 7 | 0.482 |
| Smith | 2004 | Caucasian | Breast cancer | Retrospective | 57 | 67 | 20 | 79 | 101 | 44 | 0.258 |
| Vasku | 2004 | Caucasian | Lymphoma | Retrospective | 19 | 35 | 9 | 36 | 46 | 23 | 0.259 |
| Basturk | 2005 | Caucasian | Renal cell
carcinoma | Retrospective | 15 | 10 | 0 | 27 | 13 | 9 | 0.007 |
| Campa | 2005 | Caucasian | Lung cancer | Retrospective | 629 | 954 | 412 | 615 | 993 | 374 | 0.448 |
| Cordano | 2005 | Caucasian | Lymphoma | Retrospective | 134 | 197 | 77 | 106 | 184 | 59 | 0.167 |
| Festa | 2005 | Caucasian | Basal cell
carcinoma | Retrospective | 57 | 126 | 58 | 62 | 130 | 68 | 0.993 |
| Hefler | 2005 | Caucasian | Breast cancer | Retrospective | 78 | 139 | 52 | 91 | 105 | 31 | 0.935 |
| Mazur | 2005 | Caucasian | Multiple
myeloma | Retrospective | 11 | 31 | 12 | 16 | 28 | 6 | 0.239 |
| Seifart | 2005 | Caucasian | Lung cancer | Retrospective | 47 | 52 | 17 | 90 | 107 | 46 | 0.163 |
|
Balasubramanian | 2006 | Caucasian | Breast cancer | Retrospective | 170 | 244 | 83 | 168 | 235 | 87 | 0.759 |
| Gonzalez-Zuloeta
Ladd | 2006 | Caucasian | Breast cancer | Prospective | 55 | 86 | 30 | 1,286 | 1,733 | 632 | 0.246 |
| Gunter | 2006 | Caucasian | Colorectal
cancer | Retrospective | 79 | 90 | 35 | 83 | 81 | 26 | 0.385 |
| Kamangar | 2006 | Caucasian | Gastric cancer | Prospective | 21 | 54 | 27 | 51 | 58 | 43 | 0.004 |
| Lan | 2006 | Caucasian | Lymphoma | Retrospective | 211 | 231 | 68 | 241 | 264 | 85 | 0.358 |
| Michaud | 2006 | Caucasian | Prostate
cancer | Prospective | 170 | 223 | 91 | 230 | 293 | 90 | 0.832 |
| Morgan | 2006 | Caucasian | Aneurysm | Retrospective | 40 | 40 | 6 | 867 | 1,358 | 495 | 0.360 |
| Nogueira de
Souza | 2006 | Caucasian | Cervical
cancer | Retrospective | 24 | 32 | 0 | 148 | 102 | 3 | 0.001 |
| Rothman | 2006 | Caucasian | Lymphoma | Retrospective | 1,277 | 1,658 | 564 | 1,097 | 1,470 | 499 | 0.860 |
| Theodoropoulos | 2006 | Caucasian | Colorectal
cancer | Retrospective | 111 | 76 | 35 | 64 | 86 | 50 | 0.055 |
| Vogel | 2006 | Caucasian | Breast cancer | Prospective | 108 | 167 | 86 | 98 | 177 | 86 | 0.728 |
| Wang | 2006 | Caucasian | Lymphoma | Retrospective | 486 | 474 | 174 | 393 | 410 | 138 | 0.068 |
| Berkovic | 2007 | Caucasian | GEP-NETs | Retrospective | 25 | 44 | 11 | 69 | 75 | 18 | 0.724 |
| Brenner | 2007 | Caucasian | Glioma | Retrospective | 222 | 332 | 100 | 319 | 503 | 211 | 0.621 |
| Deans | 2007 | Caucasian | Gastro-oesophageal
cancer | Retrospective | 71 | 83 | 43 | 79 | 101 | 44 | 0.258 |
| Duch | 2007 | Caucasian | Multiple
myeloma | Retrospective | 28 | 22 | 2 | 35 | 23 | 2 | 0.442 |
| Gatti | 2007 | Caucasian | Gastric cancer | Retrospective | 42 | 13 | 1 | 23 | 27 | 6 | 0.642 |
| Gonullu | 2007 | Caucasian | Breast cancer | Retrospective | 15 | 17 | 6 | 14 | 3 | 7 | <0.001 |
| Litovkin | 2007 | Caucasian | Breast cancer and
uterine leiomyoma | Retrospective | 44 | 64 | 25 | 30 | 39 | 9 | 0.490 |
| Oliveira | 2007 | Caucasian | Osteosarcoma | Retrospective | 9 | 23 | 32 | 10 | 68 | 82 | 0.405 |
| Purdue | 2007 | Caucasian | Lymphoma | Retrospective | 177 | 245 | 91 | 157 | 210 | 90 | 0.194 |
| Slattery | 2007 | Caucasian | Colorectal
cancer | Retrospective | 952 | 1,043 | 355 | 728 | 897 | 347 | 0.015 |
| Talseth | 2007 | Caucasian | Colorectal
cancer | Retrospective | 36 | 58 | 24 | 25 | 53 | 22 | 0.542 |
| Vogel | 2007 | Caucasian | Basal cell
carcinoma | Prospective | 65 | 176 | 63 | 89 | 157 | 69 | 0.988 |
| Vogel | 2007 | Caucasian | Colorectal
cancer | Prospective | 98 | 168 | 89 | 204 | 364 | 185 | 0.371 |
| Zanke | 2007 | Caucasian | Colorectal
cancer | Retrospective | 381 | 557 | 195 | 373 | 539 | 213 | 0.461 |
| Colakogullari | 2008 | Caucasian | Lung cancer | Retrospective | 10 | 29 | 5 | 27 | 22 | 9 | 0.222 |
| Crusius | 2008 | Caucasian | Gastric cancer | Prospective | 78 | 122 | 43 | 415 | 517 | 206 | 0.044 |
| Ennas | 2008 | Caucasian | Leukemia | Retrospective | 17 | 16 | 6 | 64 | 43 | 5 | 0.506 |
| Fontanella | 2008 | Caucasian | Aneurysm | Retrospective | 144 | 157 | 34 | 66 | 71 | 19 | 0.989 |
| Gu | 2008 | Caucasian | Melanoma | Prospective | 69 | 106 | 32 | 69 | 102 | 33 | 0.646 |
| Kury | 2008 | Caucasian | Colorectal
cancer | Retrospective | 363 | 489 | 171 | 435 | 504 | 182 | 0.079 |
| Vairaktaris | 2008 | Caucasian | Oral cancr | Retrospective | 42 | 102 | 18 | 90 | 60 | 6 | 0.298 |
| Vogel | 2008 | Caucasian | Lung cancer | Prospective | 105 | 202 | 96 | 204 | 361 | 179 | 0.437 |
| Wilkening | 2008 | Caucasian | Colorectal
cancer | Prospective | 79 | 163 | 61 | 162 | 297 | 121 | 0.481 |
| Aladzsity | 2009 | Caucasian | Multiple
myeloma | Retrospective | 37 | 43 | 17 | 36 | 49 | 14 | 0.681 |
| Birmann | 2009 | Caucasian | Multiple
myeloma | Prospective | 21 | 46 | 10 | 52 | 82 | 28 | 0.655 |
| Cherel | 2009 | Caucasian | Breast cancer | Retrospective | 102 | 131 | 60 | 29 | 58 | 25 | 0.695 |
| Moore | 2009 | Caucasian | Prostate
cancer | Prospective | 191 | 485 | 281 | 196 | 401 | 250 | 0.152 |
| Ozgen | 2009 | Caucasian | Papillary thyroid
carcinoma | Retrospective | 21 | 14 | 7 | 143 | 171 | 26 | 0.009 |
| Pierce | 2009 | Caucasian | Prostate
cancer | Prospective | 82 | 101 | 32 | 864 | 848 | 306 | 0.000 |
|
Talar-Wojnarowska | 2009 | Caucasian | Pancreatic
cancer | Retrospective | 13 | 19 | 9 | 22 | 19 | 9 | 0.191 |
| Tsilidis | 2009 | Caucasian | Colorectal
cancer | Prospective | 68 | 93 | 39 | 113 | 170 | 71 | 0.627 |
| Vasku | 2009 | Caucasian | Colorectal
cancer | Retrospective | 22 | 46 | 32 | 22 | 47 | 31 | 0.601 |
| Wang | 2009 | Caucasian | Prostate
cancer | Prospective | 91 | 116 | 43 | 84 | 128 | 40 | 0.448 |
| Cacev | 2010 | Caucasian | Colorectal
cancer | Retrospective | 64 | 70 | 26 | 68 | 75 | 17 | 0.582 |
| Guey | 2010 | Caucasian | Bladder cancer | Retrospective | 470 | 438 | 109 | 450 | 495 | 120 | 0.356 |
| Jakubowska | 2010 | Caucasian | Breast and ovarian
cancer | Retrospective | 135 | 227 | 102 | 73 | 144 | 73 | 0.907 |
| MARIE-GENICA | 2010 | Caucasian | Breast cancer | Retrospective | 986 | 1,571 | 585 | 1,774 | 2,671 | 1,036 | 0.586 |
| Consortium | | | | | | | | | | | |
| Schonfeld | 2010 | Caucasian | Breast cancer | Retrospective | 274 | 408 | 156 | 379 | 487 | 211 | 0.017 |
| Giannitrapani | 2011 | Caucasian | Hepatocellular
carcinoma | Retrospective | 63 | 36 | 6 | 51 | 37 | 10 | 0.402 |
| Grimm | 2011 | Caucasian | Cervical
cancer | Retrospective | 55 | 51 | 25 | 85 | 96 | 27 | 0.990 |
| Bushley | 2004 | Mixed | Ovarian cancer | Retrospective | 5 | 34 | 143 | 9 | 46 | 163 | 0.020 |
| Dossus | 2010 | Mixed | Prostate
cancer | Prospective | 3,594 | 3,218 | 1,125 | 3,832 | 3,402 | 1,274 | <0.001 |
| Dossus | 2010 | Mixed | Breast cancer | Prospective | 2,847 | 2,523 | 820 | 3,707 | 3,324 | 1,035 | <0.001 |
| Ognjanovic | 2010 | Mixed | Colorectal
cancer | Retrospective | 173 | 74 | 22 | 357 | 136 | 43 | <0.001 |
Test of heterogeneity
There was a significant heterogeneity in overall
comparison of the CC genotype vs. GG genotype (P<0.001 and
I2=43.8%). The meta-regression showed that the strong
heterogeneity could not be traditionally explained by cancer types,
ethnicities or study designs (P=0.285, 0.129 and 0.306,
respectively). Furthermore, the 15 studies that deviate from HWE
showed similar heterogeneity with that of studies that were in HWE
(Table II), suggesting that the
remarkable heterogeneity among the overall analysis was not due to
the variability of the control quality. Therefore, we carefully
assessed the association of the IL6 -174G/C polymorphism
with cancer risk in several subgroups, and carried out sensitivity
analyses of between-study heterogeneity to detect studies that have
remarkable influence on homogeneity.
 | Table II.Summary of ORs and 95% CIs for the
IL6 -174G>C polymorphism and cancer risk and
heterogeneity test for studies of each group. |
Table II.
Summary of ORs and 95% CIs for the
IL6 -174G>C polymorphism and cancer risk and
heterogeneity test for studies of each group.
| Variables | No.a | Cases/controls | CC vs. GG
| GC vs. GG
| GC/CC vs. GG
|
|---|
| | | OR (95% CI)b |
PHc | I2
(%) | OR (95% CI)b |
PHc | I2
(%) | OR (95% CI)b |
PHc | I2
(%) |
|---|
| Total | 84 | 44,735/60,747 | 1.01
(0.95–1.08) | <0.001 | 43.8 | 1.01
(0.96–1.07) | <0.001 | 52.5 | 1.01
(0.96–1.07) | <0.001 | 57.0 |
| Ethnicities | | | | | | | | | | | |
| African | 7 | 1,138/1,299 | 1.83
(1.26–2.67)e | 0.294 | 17.7 | 0.80
(0.55–1.16) | 0.002 | 71.0 | 0.94
(0.67–1.31) | 0.004 | 68.9 |
| Caucasian | 73 | 29019/42120 | 1.00
(0.92–1.08) | <0.001 | 43.8 | 1.02
(0.96–1.09) | <0.001 | 52.4 | 1.02
(0.96–1.08) | <0.001 | 58.2 |
| Mixed | 4 | 14,578/17,328 | 0.98
(0.92–1.05) | 0.488 | 0.0 | 1.00
(0.96–1.05) | 0.839 | 0.0 | 1.00
(0.95–1.04) | 0.789 | 0.0 |
| Cancer
typesd | | | | | | | | | | | |
| Breast
cancer | 13 | 12,640/20,281 | 1.01
(0.94–1.09) | 0.374 | 7.2 | 1.06
(0.96–1.17) | 0.033 | 46.4 | 1.05
(0.95–1.15) | 0.043 | 44.3 |
| Colorectal
cancer | 13 | 6,798/7,502 | 0.97
(0.82–1.14) | 0.014 | 52.4 | 1.01
(0.89–1.13) | 0.030 | 47.2 | 1.00
(0.87–1.13) | 0.002 | 61.9 |
| Prostate
cancer | 6 | 10,043/12,438 | 0.99
(0.91–1.07) | 0.254 | 24.0 | 1.03
(0.97–1.09) | 0.361 | 8.6 | 1.01
(0.96–1.07) | 0.357 | 9.3 |
| Lymphomad | 7 | 6,213/5,586 | 0.96
(0.86–1.07) | 0.574 | 0.0 | 0.96
(0.89–1.04) | 0.609 | 0.0 | 0.96
(0.89–1.03) | 0.621 | 0.0 |
| Multiple
myelomad | 6 | 422/601 | 1.23
(0.82–1.83) | 0.609 | 0.0 | 1.07
(0.80–1.41) | 0.717 | 0.0 | 1.10
(0.84–1.44) | 0.731 | 0.0 |
| Lung
cancerd | 5 | 2,801/3,234 | 1.06
(0.92–1.23) | 0.715 | 0.0 | 1.05
(0.83–1.33) | 0.071 | 53.7 | 1.01
(0.90–1.13) | 0.153 | 40.2 |
| Gastric
cancerd | 5 | 554 /1,585 | 1.06
(0.78–1.44) | 0.114 | 46.2 | 0.98
(0.55–1.73) | 0.001 | 78.8 | 0.95
(0.54–1.76) | <0.001 | 80.4 |
| Melanomad | 3 | 410/476 | 1.13
(0.75–1.68) | 0.790 | 0.0 | 1.18
(0.88–1.58) | 0.721 | 0.0 | 1.17
(0.88–1.54) | 0.672 | 0.0 |
| Cervical
cancer | 3 | 347/661 | 1.85
(1.12–3.08)f | 0.320 | 12.2 | 1.11
(0.68–1.82) | 0.068 | 62.7 | 1.22
(0.92–1.61) | 0.195 | 38.7 |
| Other types | 28 | 4,723/9,403 | 1.10
(0.85–1.43) | <0.001 | 67.6 | 0.94
(0.79–1.11) | <0.001 | 67.1 | 0.97
(0.82–1.16) | <0.001 | 71.5 |
| Study design | | | | | | | | | | | |
| Prospective | 17 | 18,759/28,718 | 1.01
(0.95–1.07) | 0.893 | 0.0 | 1.03
(0.98–1.07) | 0.150 | 26.6 | 1.02
(0.98–1.06) | 0.316 | 11.8 |
|
Retrospective | 67 | 25,976/32,029 | 1.00
(0.91–1.11) | <0.001 | 51.9 | 0.98
(0.92–1.06) | <0.001 | 56.3 | 0.99
(0.92–1.06) | <0.001 | 61.9 |
| HWE | | | | | | | | | | | |
| >0.05 | 69 | 26,067/36,098 | 1.01
(0.93–1.09) | <0.001 | 44.7 | 1.00
(0.94–1.07) | <0.001 | 50.8 | 1.00
(0.94–1.07) | <0.001 | 59.0 |
| <0.05 | 15 | 18,668/24,649 | 1.02
(0.90–1.15) | 0.043 | 42.3 | 1.06
(0.95–1.18) | 0.001 | 61.4 | 1.04
(0.96–1.13) | 0.019 | 48.3 |
For ethnic-specific subgroup analysis, no
heterogeneity was detected within African population studies
(P=0.294), but there were significant heterogeneity within
Caucasian population studies (P<0.001). Sensitivity analysis of
between-study heterogeneity revealed that five studies (30–34)
mainly contributed to the heterogeneity within Caucasian population
studies. After performing cancer type-specific analyses, we found
no heterogeneity in studies of breast, prostate, lung, gastric
cancer, lymphoma, multiple myeloma, melanoma and cervical cancer
(Table II). However, there was
strong heterogeneity for colorectal cancer, which was due to one
Caucasian study (32).
Quantitative data synthesis
Overall, the CC genotype was not significantly
associated with cancer risk when compared to the GG genotype
(OR=1.01, 95% CI 0.95–1.08, P=0.698). Ethnic-specific ORs showed
that cancer risk was increased for individuals carrying the CC
genotype compared to those with the GG genotype in African
populations (OR=1.83, 95% CI 1.26–2.67, P=0.002; Fig. 2), but not in Caucasian populations
(OR=1.00, 95% CI 0.92–1.08, P=0.938; Table II). After excluding the studies
(30–34) responsible for heterogeneity, we
found that Caucasian individuals carrying the CC genotype had no
remarkable effect on risk of cancer compared to GG genotype
individuals (OR=1.02, 95% CI 0.97–1.07, P=0.561) with no
significant between-study heterogeneities (P=0.196,
I2=12.6%). Although there were nine data sets in which
the genotype distribution did not follow HWE, the corresponding
meta-analysis was qualitatively similar with or without excluding
them.
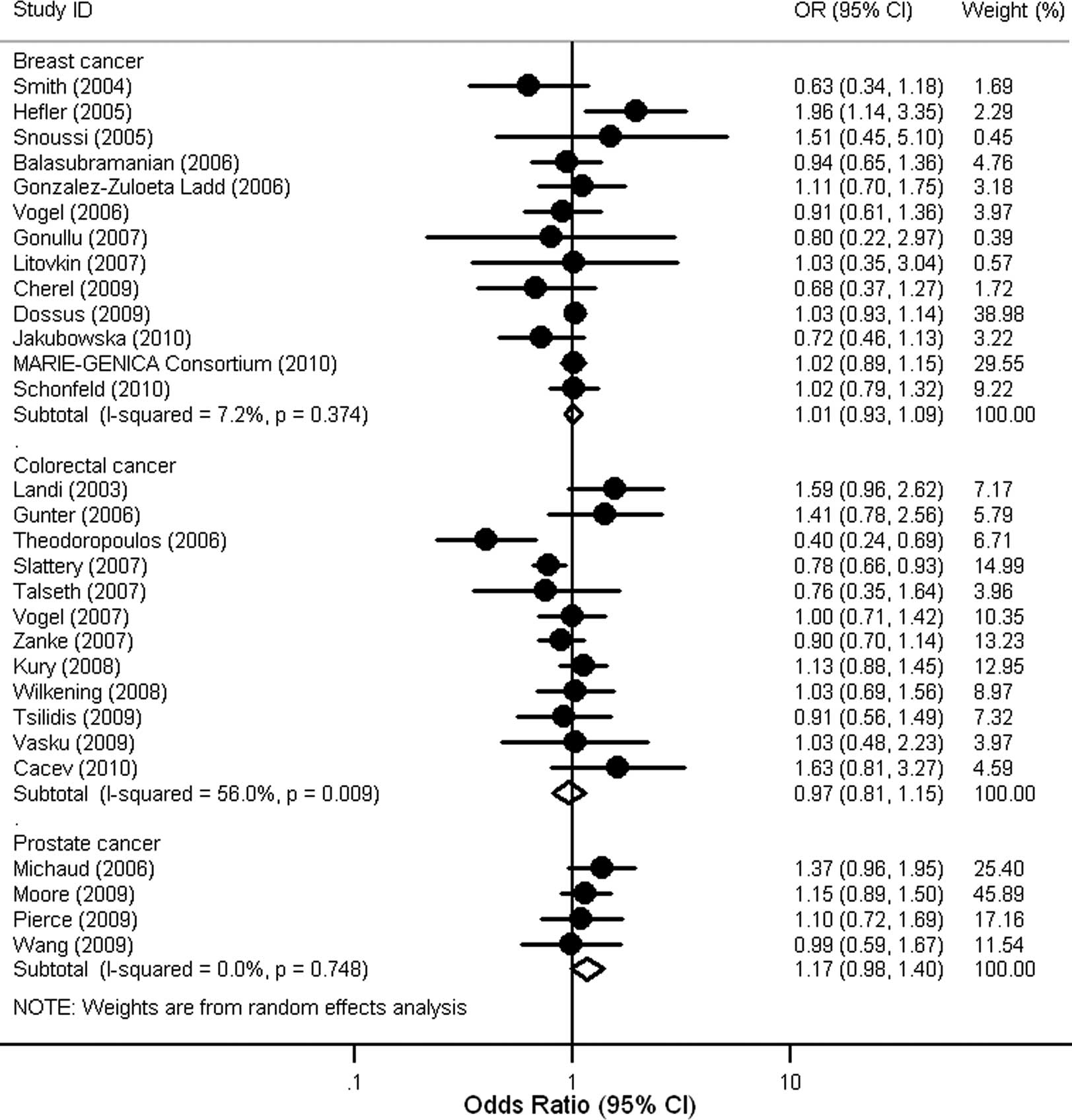
Subsequently, we stratified the association between
the IL6 -174G/C polymorphism and cancer risk by cancer
types. When compared to individuals with the GG genotype, those
with the CC genotype were associated with increased risk of
cervical cancer (OR=1.85, 95% CI 1.12–3.08, P=0.017), but not with
that of other types of cancer (Table
II). Furthermore, no significant association of the IL6
-174G/C polymorphism with risk of breast, colorectal and prostate
cancer was observed in individuals with Caucasian ancestry
(Fig. 3). A few studies involving
African populations did not allow us to perform subgroup analysis
in Africans (Table I).
Lastly, we also assessed the ORs of cancer for
individuals with the GC genotype or C allele carriers (GC or CC
genotype) compared to those with the GG genotype, and found no
significant association in overall and subgroup analyses (Table II).
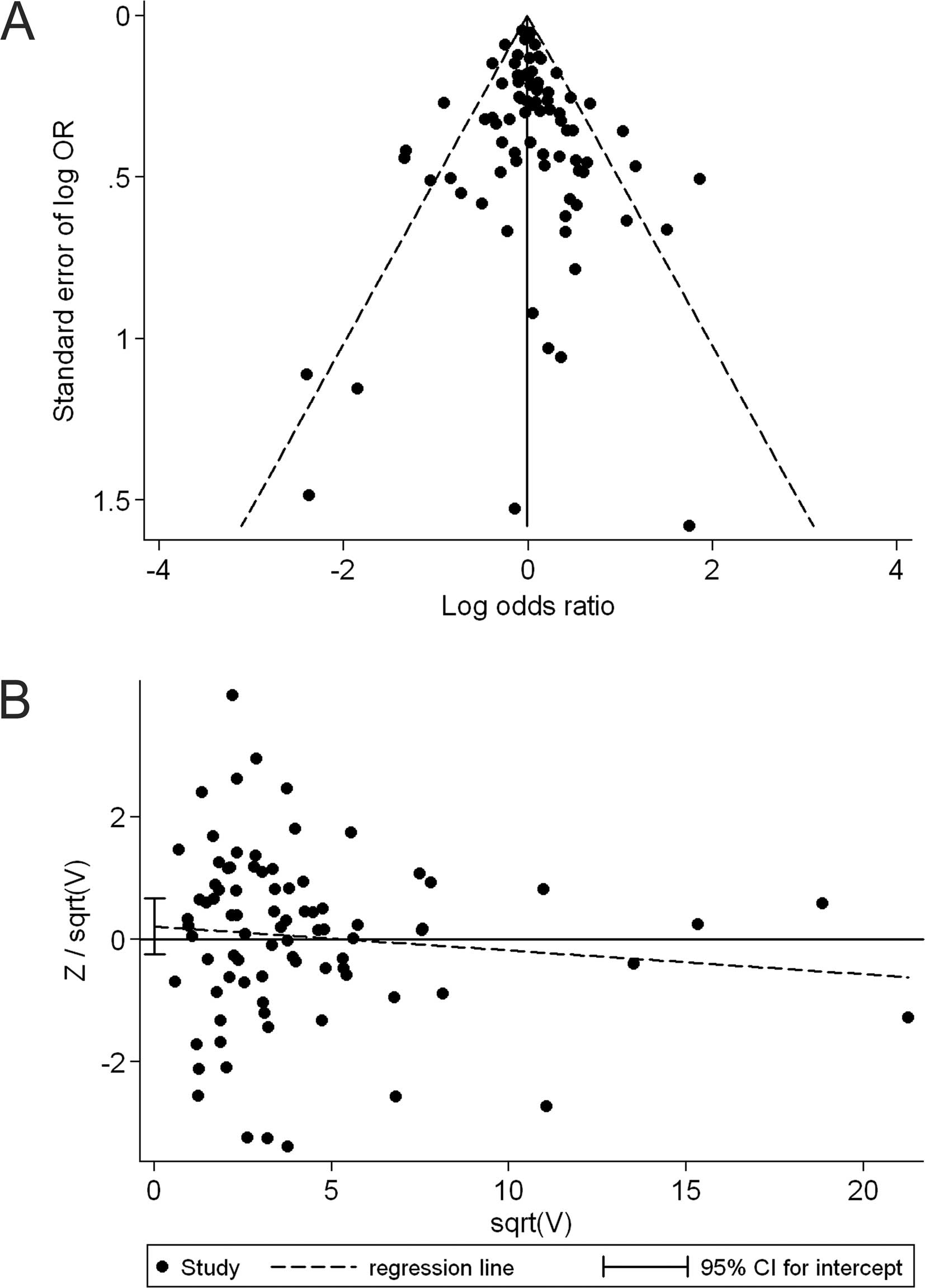
Publication bias
The shape of the funnel plot did not reveal any
evidence of obvious asymmetry in comparison of the CC genotype vs.
GG genotype (Fig. 4A). Then, we
used Harbord's test to provide statistical evidence of funnel plot
symmetry, and the result did not show evidence of publication bias
(t=0.91, P=0.366; Fig. 4B).
Subgroup analyses by ethnicity and cancer type did not provide any
evidence of publication bias (t=−1.59, P=0.172 for African
populations; t=0.27, P=0.786 for Caucasian populations; t=−0.58,
P=0.575 for breast cancer and t=1.02, P=0.329 for colorectal
cancer). Similarly, no publication bias was detected when comparing
the GC genotype to the GG genotype (t=0.18, P=0.861), the GC/CC
genotype to the GG genotype (t=0.42, P=0.677) and in any comparison
of the corresponding subgroup analyses.
Discussion
To the best of our knowledge, the present
meta-analysis of 83 studies, involving 44,735 cases and 60,747
controls (counting every study's cases and controls only once),
provides the most comprehensive assessment of an association of the
IL6 -174G/C polymorphism with cancer risk. It provides
evidence that African individuals with the CC genotype have higher
odds of cancer than individuals with the GG genotype; the findings
of our meta-analysis do not show any association of the IL6
-174G/C with cancer risk in Caucasian populations. These findings
suggest an ethnic-specific effect of IL6 -174G/C
polymorphism on risk of cancer. The discrepancies among different
populations suggest a possible role of ethnic differences in
genetic backgrounds and the environment they lived in (35).
Recent studies have shown that IL6 and its major
effector STAT3 play a role in the epigenetic switch from
non-transformed epithelia to cancer cell (36,37).
Elevated expression of IL6 via autocrine and paracrine mechanisms
leading to subsequent chronic inflammation also exhibits a strong
association with cancer (38–41).
In the present study, we found that the IL6 -174 CC genotype
was significantly associated with increased risk of cervical cancer
compared to the GG genotype. However, the smaller number of
individuals genotyped in these studies precludes any formal
conclusion. As compared to previous analyses based on substantially
less data (15,16), the present analysis essentially
shows null associations between IL6 -174G/C and several
common types of cancer, including breast, colorectal, prostate,
lung, gastric cancer, lymphoma, multiple myeloma and melanoma.
Assessment of the between-study heterogeneity and
identification of its sources are essential requirements in
meta-analyses (23,42). In this study, we systematically
examined the effect of IL6 -174G/C on cancer risk across all
reliable studies, and the results of the overall analysis showed a
strong heterogeneous effect among the 83 studies. Given the fact
that clinical characteristics of studies, including study
population, design approach and type of cancer, are likely to be
potential sources of heterogeneity, we first used meta-regression
to detect whether any of the characteristics could explain the
between-study variation. However, none of the potential sources
considered were able to systematically explain the observed
variation across studies. It seems likely that there exist more
than one answer to the nature of overall heterogeneity. We,
therefore, induced a new approach to perform sensitivity analysis
of between-study heterogeneity (24), and detected that several studies
with different clinical characteristics were responsible for the
heterogeneity (30–34).
Apart from between-study heterogeneity, publication
bias has also been recognized as a major concern in robust
meta-analyses. Thereby, in this study, we used funnel plots to
assess whether the studies included could be affected by
publication bias. According to the recommendations by Sterne and
Egger (43), the log OR and its
standard error are used for the horizontal and vertical axis,
respectively. No evidence of publication bias was found when
testing by a visual inspection of funnel plots. In support of this,
Harbord's linear regression test confirmed the evidence of funnel
plot symmetry across all constituent data sets.
Notably, social factors are believed to interact
with genetic variants to govern complex human phenotypes (44,45).
Cole et al recently demonstrated a strong interaction
between the IL6 -174G/C polymorphism and social environment
factors, which may further affect the risk of inflammation-related
disease (46). However, in the
absence of the original data of the reviewed studies, our
evaluation of potential interactions of gene-environment with
cancer risk was limited. This may explain why previous genetic
association studies and some subgroup analyses in our
meta-analysis, especially the Caucasian studies, failed to show an
association between the IL6 -174G/C polymorphism and risk of
cancer. Furthermore, two other polymorphisms (-6331T/C and -572G/C)
and several haplotypes in the IL6 promoter affect the
transcriptional activity of IL6 and may influence
susceptibility to inflammation-related diseases (47–49).
However, most studies included in our meta-analysis restricted
their analysis to the IL6 -174G/C polymorphism and few
carried out the IL6 haplotypic analysis on cancer
susceptibility. It is difficult to estimate the role of a
particular haplotype on cancer risk in the present
meta-analysis.
Despite these limitations, our meta-analysis
provides a leap in knowledge when compared to a previous study
(15) that reviewed the
association between the IL6 -174G/C polymorphism and risk of
cancer. First, our updated review is more comprehensive than the
previous, as we identified 83 independent articles with a total of
105,482 individuals on the association of IL6 -174G/C with
cancer risk compared to 47 articles with 67,116 individuals in the
previous report. Thus, our meta-analysis had significantly higher
statistical power. Second, we noticed the potentially different
roles of the IL6 -174G/C polymorphism in the development of
cancer among various populations, and found different associations
of this polymorphism with cancer risk between Africans and
Caucasians. Third, sensitivity analysis of heterogeneity was used
to detect studies that were responsible for between-study
heterogeneity (24). Fourth, we
assessed the pooled effect of the IL6 -174G/C polymorphism
on cancer risk within or without the studies that did not follow
HWE, and qualitatively similar results were found, suggesting that
the estimations of our analyses are stable and convincing. Finally,
for publication bias analysis, a modified method for testing funnel
plot asymmetry was used (28),
which maintains better control of the false-positive rate than the
commonly used Egger's test. No publication bias was detected,
suggesting that the pooled results should be unbiased.
In summary, the present meta-analysis provides
evidence of the ethnic-specific association of the IL6
-174G/C polymorphism with cancer risk. More sophisticated
gene-environment interactions should be considered in future
analyses, which may result in better understanding of the relevance
between the IL6 -174G/C polymorphism and risk of cancer.
Moreover, this study reinforces the need to undertake
investigations with very large number of cases and controls
(including updated meta-analyses) to provide conclusive evidence
for the associations between high-frequency genetic variants in
low-penetrance genes and complex diseases, such as cancer.
Acknowledgements
This study was, in part, supported by
grants from the National Natural Science Foundation of China
(81171894, 30973425 and 30672400 to H.T. Zhang), the Program for
New Century Excellent Talents in University (NCET-09-0165 to H.T.
Zhang), the Science and Technology Committee of Jiangsu Province
(BK2008162 to H.T. Zhang), the SRF for ROCS, State Education
Ministry (2008890 to H.T. Zhang), the Qing-Lan Project of Education
Bureau of Jiangsu Province (to H.T. Zhang), the ‘333’ Project of
Jiangsu Province Government (to H.T. Zhang), and the Soochow
Scholar Project of Soochow University (to H.T. Zhang). This study
was also supported by grants #2R01GM069430-06, CA 137000, CA 112520
and CA 108741 from the NIH (to B. Pasche).
References
|
1.
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar
|
|
3.
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar
|
|
4.
|
Balkwill F and Mantovani A: Inflammation
and cancer: back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Engels EA, Wu X, Gu J, Dong Q, Liu J and
Spitz MR: Systematic evaluation of genetic variants in the
inflammation pathway and risk of lung cancer. Cancer Res.
67:6520–6527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Tindall EA, Severi G, Hoang HN, et al:
Comprehensive analysis of the cytokine-rich chromosome 5q31.1
region suggests a role for IL-4 gene variants in prostate cancer
risk. Carcinogenesis. 31:1748–1754. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kishimoto T: Interleukin-6: from basic
science to medicine – 40 years in immunology. Annu Rev Immunol.
23:1–21. 2005.
|
|
8.
|
Katsumata N, Eguchi K, Fukuda M, et al:
Serum levels of cytokines in patients with untreated primary lung
cancer. Clin Cancer Res. 2:553–559. 1996.PubMed/NCBI
|
|
9.
|
Bromberg JF, Wrzeszczynska MH, Devgan G,
et al: Stat3 as an oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar
|
|
10.
|
Grivennikov S, Karin E, Terzic J, et al:
IL-6 and Stat3 are required for survival of intestinal epithelial
cells and development of colitis-associated cancer. Cancer Cell.
15:103–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Walter M, Liang S, Ghosh S, Hornsby PJ and
Li R: Interleukin 6 secreted from adipose stromal cells promotes
migration and invasion of breast cancer cells. Oncogene.
28:2745–2755. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Sullivan NJ, Sasser AK, Axel AE, et al:
Interleukin-6 induces an epithelial-mesenchymal transition
phenotype in human breast cancer cells. Oncogene. 28:2940–2947.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Fishman D, Faulds G, Jeffery R, et al: The
effect of novel polymorphisms in the interleukin-6 (IL-6) gene on
IL-6 transcription and plasma IL-6 levels, and an association with
systemic-onset juvenile chronic arthritis. J Clin Invest.
102:1369–1376. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Belluco C, Olivieri F, Bonafe M, et al:
-174 G>C polymorphism of interleukin 6 gene promoter affects
interleukin 6 serum level in patients with colorectal cancer. Clin
Cancer Res. 9:2173–2176. 2003.
|
|
15.
|
Xu B, Niu XB, Wang ZD, et al: IL-6
-174G>C polymorphism and cancer risk: a meta-analysis involving
29,377 cases and 37,739 controls. Mol Biol Rep. 38:2589–2596.
2011.
|
|
16.
|
Yu KD, Di GH, Fan L, Chen AX, Yang C and
Shao ZM: Lack of an association between a functional polymorphism
in the interleukin-6 gene promoter and breast cancer risk: a
meta-analysis involving 25,703 subjects. Breast Cancer Res Treat.
122:483–488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: Preferred reporting items for systematic reviews and
meta-analyses: the PRISMA statement. BMJ. 339:b25352009. View Article : Google Scholar
|
|
18.
|
Stroup DF, Berlin JA, Morton SC, et al:
Meta-analysis of observational studies in epidemiology: a proposal
for reporting. Meta-analysis of Observational Studies in
Epidemiology (MOOSE) group. JAMA. 283:2008–2012. 2000. View Article : Google Scholar
|
|
19.
|
Yu W, Gwinn M, Clyne M, Yesupriya A and
Khoury MJ: A navigator for human genome epidemiology. Nat Genet.
40:124–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Cochran WG: The combination of estimates
from different experiments. Biometrics. 10:101–129. 1954.
View Article : Google Scholar
|
|
21.
|
Dickersin K and Berlin JA: Meta-analysis:
state-of-the-science. Epidemiol Rev. 14:154–176. 1992.
|
|
22.
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lau J, Ioannidis JP and Schmid CH: Summing
up evidence: one answer is not always enough. Lancet. 351:123–127.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Patsopoulos NA, Evangelou E and Ioannidis
JP: Sensitivity of between-study heterogeneity in meta-analysis:
proposed metrics and empirical evaluation. Int J Epidemiol.
37:1148–1157. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
26.
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Harbord RM, Egger M and Sterne JA: A
modified test for small-study effects in meta-analyses of
controlled trials with binary endpoints. Stat Med. 25:3443–3457.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Dossus L, Kaaks R, Canzian F, et al: PTGS2
and IL6 genetic variation and risk of breast and prostate cancer:
results from the Breast and Prostate Cancer Cohort Consortium
(BPC3). Carcinogenesis. 31:455–461. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Foster CB, Lehrnbecher T, Samuels S, et
al: An IL6 promoter polymorphism is associated with a lifetime risk
of development of Kaposi sarcoma in men infected with human
immunodeficiency virus. Blood. 96:2562–2567. 2000.PubMed/NCBI
|
|
31.
|
Morgan L, Cooper J, Montgomery H, Kitchen
N and Humphries SE: The interleukin-6 gene -174G>C and
-572G>C promoter polymorphisms are related to cerebral
aneurysms. J Neurol Neurosurg Psychiatry. 77:915–917. 2006.
|
|
32.
|
Theodoropoulos G, Papaconstantinou I,
Felekouras E, et al: Relation between common polymorphisms in genes
related to inflammatory response and colorectal cancer. World J
Gastroenterol. 12:5037–5043. 2006.PubMed/NCBI
|
|
33.
|
Vairaktaris E, Yapijakis C, Serefoglou Z,
et al: Gene expression polymorphisms of interleukins-1 beta, -4,
-6, -8, -10, and tumor necrosis factors-alpha, -beta: regression
analysis of their effect upon oral squamous cell carcinoma. J
Cancer Res Clin Oncol. 134:821–832. 2008. View Article : Google Scholar
|
|
34.
|
Slattery ML, Wolff RK, Herrick JS, Caan BJ
and Potter JD: IL6 genotypes and colon and rectal cancer. Cancer
Causes Control. 18:1095–1105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Hirschhorn JN, Lohmueller K, Byrne E and
Hirschhorn K: A comprehensive review of genetic association
studies. Genet Med. 4:45–61. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Iliopoulos D, Hirsch HA and Struhl K: An
epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and
IL6 links inflammation to cell transformation. Cell. 139:693–706.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk
ML and Struhl K: STAT3 activation of miR-21 and miR-181b-1 via PTEN
and CYLD are part of the epigenetic switch linking inflammation to
cancer. Mol Cell. 39:493–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Okamoto M, Lee C and Oyasu R:
Interleukin-6 as a paracrine and autocrine growth factor in human
prostatic carcinoma cells in vitro. Cancer Res. 57:141–146.
1997.PubMed/NCBI
|
|
39.
|
Yeh HH, Lai WW, Chen HH, Liu HS and Su WC:
Autocrine IL-6-induced Stat3 activation contributes to the
pathogenesis of lung adenocarcinoma and malignant pleural effusion.
Oncogene. 25:4300–4309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Bromberg J and Wang TC: Inflammation and
cancer: IL-6 and STAT3 complete the link. Cancer Cell. 15:79–80.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Park EJ, Lee JH, Yu GY, et al: Dietary and
genetic obesity promote liver inflammation and tumorigenesis by
enhancing IL-6 and TNF expression. Cell. 140:197–208. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Thompson SG: Why sources of heterogeneity
in meta-analysis should be investigated. BMJ. 309:1351–1355.
1994.PubMed/NCBI
|
|
43.
|
Sterne JA and Egger M: Funnel plots for
detecting bias in meta-analysis: guidelines on choice of axis. J
Clin Epidemiol. 54:1046–1055. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Robinson GE: Genomics. Beyond nature and
nurture Science. 304:397–399. 2004.
|
|
45.
|
Frazer KA, Murray SS, Schork NJ and Topol
EJ: Human genetic variation and its contribution to complex traits.
Nat Rev Genet. 10:241–251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Cole SW, Arevalo JM, Takahashi R, et al:
Computational identification of gene-social environment interaction
at the human IL6 locus. Proc Natl Acad Sci USA. 107:5681–5686.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Gu W, Du DY, Huang J, et al:
Identification of interleukin-6 promoter polymorphisms in the
Chinese Han population and their functional significance. Crit Care
Med. 36:1437–1443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Smith AJ, D'Aiuto F, Palmen J, et al:
Association of serum interleukin-6 concentration with a functional
IL6 -6331T>C polymorphism. Clin Chem. 54:841–850.
2008.PubMed/NCBI
|
|
49.
|
Terry CF, Loukaci V and Green FR:
Cooperative influence of genetic polymorphisms on interleukin 6
transcriptional regulation. J Biol Chem. 275:18138–18144. 2000.
View Article : Google Scholar : PubMed/NCBI
|