Introduction
Oral squamous cell carcinoma is one of the most
common cancers in the world and its main risk factors are cigarette
smoking and alcohol consumption (1,2) as
well as betel quid chewing (3).
Increasing epidemiological evidence suggests their complex
interactions between numerous genetic and environmental factors are
major causes. Although individuals are exposed to environmental
risk factors and extensive tobacco, alcohol consumption and betel
quid chewing, oral cancer develops only in a small proportion of
exposed individuals, implying that genetic factors may play a role
in its carcinogenic mechanisms.
Our previous meta-analyses revealed GSTM1 genetic
variation as a risk factor for oral cancer, whereas the CYP1A1
(4) or TP53 (5) polymorphism may not increase the
susceptibility to oral neoplasm. In addition, meta-analyses
conducted in 2009 and 2011, respectively, suggested XRCC1 (6) and CYP2E1 (7) polymorphisms as biomarkers for oral
carcinoma in Asian populations. However, although genetic factors
are important, only a few genes associated with genetic
susceptibility to oral cancer were identified.
Cyclin D1 (CCND1) is a key regulatory protein that
plays an important role in the transition from G1 to S phase of the
cell cycle during cell division. Inhibition of CCND1 function often
results in cell cycle arrest, whereas overexpression of CCND1 may
disrupt normal cell cycle control and subsequently contribute to
oncogenesis (8). Evidence suggests
that gene amplification and protein overexpression of CCND1 were
detected in a number of cancers, including oral cancer (9–11),
and were considered to be associated with a poor prognosis. Hence,
CCND1 has been regarded as a potential target for tumor therapy
(12,13).
Previously, a common functional polymorphism, G870A,
of CCND1 has been widely studied as a possible low-penetrant
susceptibility allele for various cancers (14). CCND1 G870A is a silent variant that
does not result in an amino acid alteration within the protein
sequence. The A allele has been considered to result in an
alternatively spliced transcript of CCND1, namely, transcript b.
This alternate transcript lacks exon 5 of CCND1 that contains a
PEST-rich region, critical for the degradation of CCND1. Thus,
transcript b (A allele) has been shown to have a longer half-life
than the transcript a (G allele, the wild-type gene) encoded
protein, resulting in different biochemical and biological protein
features (15). It has been
demonstrated that transcript b is more likely to bypass the G1-S
cell cycle checkpoint, which consequently leads to carcinogenesis
(16). As a result, three distinct
genotypes were created, namely, homozygous AA, homozygous GG and
heterozygous AG. Evidence suggests that the CCND1 polymorphism may
have a correlation with its overexpression in cancers (17,18).
Thus, evaluation of the association of CCND1 polymorphism with oral
cancer risk is required.
Published data on the possible association of CCND1
G870A polymorphism with oral carcinoma have generated inconclusive
results. To the best of our knowledge, the question of whether the
CCND1 G870A polymorphism increases oral cancer risk remains
uncertain. To clarify this association may elucidate the possible
risk of oral cancer and therefore contribute to its prevention. As
a single study may have been underpowered in clarifying the
associations of CCND1 G870A polymorphisms with oral carcinoma
susceptibility, in the present study we performed evidence-based
quantitative meta-analyses that increased statistical power to
address this controversy.
Materials and methods
Literature search strategy
We carried out a search in the Medline, EMBASE,
OVID, Sciencedirect and CNKI databases without a language
limitation, including all manuscripts published until February
2012, with a combination of the following keywords: cyclin D1,
CCND1, oral, mouth neoplasm, tumor, cancer, variation and
polymorphism. All searched studies were retrieved and the
bibliographies were checked for other relevant publications. Review
articles and bibliographies of other relevant studies identified
were hand-searched to find additional eligible studies.
Inclusion criteria
The following criteria were used for the literature
selection: First, studies should concern the association of CCND1
G870A polymorphism with oral cancer risk; second, studies should be
observational studies (case-control or cohort); third, manuscripts
must offer the size of the sample, odds ratios (ORs) and their 95%
confidence intervals (CIs), the genetic distribution or the
information to infer the results. After searching, we reviewed all
manuscripts in accordance with the criteria defined above for
further analysis.
Data extraction
Data were extracted from all eligible publications
independently by two of the authors according to the inclusion
criteria mentioned above. For conflicting evaluations, an agreement
was reached following a discussion. If a consensus could not be
reached, another author was consulted to resolve the dispute and
then a final decision was made by the majority of the votes. The
extracted information was entered into a database. For data not
provided in the main text, the relevant information was obtained by
contacting corresponding authors if and when possible.
Statistical analysis
The OR of CCND1 G870A polymorphisms and oral cancer
was estimated for each study. The pooled ORs were calculated for
the additive model (AA vs. GG), dominant model (AA+AG vs. GG) and
recessive model (AA vs. AG+GG), respectively. For detection of any
possible sample size biases, the OR and its 95% CI from each study
was plotted against the number of participants. A Chi-square based
Q-statistic test was performed to assess heterogeneity. If the
result of the heterogeneity test was P>0.1, ORs were pooled
according to the fixed-effects model (Mantel-Haenszel), otherwise,
the random-effects model (DerSimonian-Laird) was used. The
significance of the pooled ORs was determined by Z-test. The
Hardy-Weinberg equilibrium (HWE) was assessed via Fisher’s exact
test.
Publication bias was assessed by visual inspection
of funnel plots (19), in which
the standard error of log (OR) of each study was plotted against
its log (OR). An asymmetric plot indicates a possible publication
bias. The symmetry of the funnel plot was further evaluated by
Egger’s linear regression test (20). Statistical analysis was undertaken
using the program Review Manager 5 and Stata 11.0 softwares (Stata
Corporation, College Station, TX, USA).
Results
Study characteristics
Publications relevant to the key words were
retrieved and screened. A total of 52 studies on CCND1 were
searched and screened for retrieval, of which 43 irrelevant studies
were excluded. Two studies (21,22)
were then excluded since the data were not confined to oral cancer.
One study (23) was excluded due
to it being a review and six studies (17,24–28)
were excluded since they concerned only CCND1 expression but not
its polymorphism. A total of six case-control studies (29–34)
were selected.
All the included six studies were written in
English. We established a database according to the extracted
information from each article. The relevant information is listed
in Table I. The first author and
the number and characteristics of cases and controls for each study
as well as other necessary information are presented.
 | Table ICharacteristics of the studies
included in the meta-analysis. |
Table I
Characteristics of the studies
included in the meta-analysis.
| First author | Year of
publication | No. of cases
(male/female) | No. of controls
(male/female) | Type of
controls | Age range (mean),
years
| Country |
|---|
| Cases | Controls |
|---|
| Matthias (34) | 1998 | 38 (25/13) | 191 (149/42) | 191 non-cancerous
controls (hospital-based) | NA (59.29) | NA (NA) | Germany |
| Wong (33) | 2003 | 70 (65/5) | 93 (74/19) | 93 controls
(population-based) | 31–82 (53.3) | 22–75 (49.4) | China |
| Holley (32) | 2005 | 174 (135/39) | 155 (125/30) | 155 non-cancerous
controls (hospital-based) | NA (55.6) | NA (59.9) | Germany |
| Sathyan (31) | 2006 | 176 (119/57) | 142 (98/44) | 142 non-cancerous
controls (age-, gender-matched; hospital-based) | 36–85 (59) | 35–80 (58) | India |
| Gomes (30) | 2008 | 80 (60/20) | 80 (59/21) | 80 healthy
individuals (age-, gender, tobacco usage-matched;
population-based) | 39–82 (57.5) | 39–85 (53.5) | Brazil |
| Tsai (29) | 2011 | 620 (586/34) | 620 (586/34) | 620 non-cancerous
controls (age-, gender-, habit-matched; hospital-based) | NA (52.4) | NA (51.3) | China |
Of the included studies, two studies were on
Caucasian (32,34), three on Asian (29,31,33)
and one on mixed ethnicity populations (30). Data with regard to smoking status
was available in two studies (29,32).
As shown in Table
II, the distributions of the CCND1 G870A genotype of the
included studies are also presented. The genetic distributions of
the control groups of two studies (29,30)
deviated from HWE, while the remaining studies did not.
 | Table IIDistribution of the CCND1 genotype
among oral cancer cases and controls included in the
meta-analysis. |
Table II
Distribution of the CCND1 genotype
among oral cancer cases and controls included in the
meta-analysis.
| First author | Genotyping
method | Cases
| Controls
| HWE (control)
|
|---|
| AA | AG | GG | AA | AG | GG | Chi-square | P-value |
|---|
| Matthias | PCR-RFLP | 11 | 20 | 7 | 35 | 101 | 55 | 0.918 | >0.05 |
| Wong | PCR-SSCP | 19 | 36 | 15 | 27 | 49 | 17 | 0.406 | >0.05 |
| Holley | PCR-RFLP | 14 | 94 | 66 | 28 | 87 | 40 | 2.593 | >0.05 |
| Sathyan | PCR-SSCP | 39 | 71 | 36 | 36 | 61 | 40 | 1.620 | >0.05 |
| Gomes | PCR-RFLP | 25 | 30 | 25 | 23 | 29 | 28 | 5.926 | <0.05 |
| Tsai | PCR-RFLP | 213 | 323 | 84 | 155 | 365 | 100 | 21.625 | <0.05 |
Test of heterogeneity
We analyzed the heterogeneity of an additive model
(AA vs. GG), dominant model (AA+AG vs. GG) and recessive model (AA
vs. AG+GG).
As shown in Table
III, the heterogeneity for the overall data with regard to oral
cancer was significant in additive, dominant and recessive models
since the P-values were <0.1 for Q-tests. However, subgroup
analyses with regard to ethnicity, smoking status and source of
control were conducted. The P-value for heterogeneity indicated a
reduced or removed heterogeneity when the data were divided into
relevant subgroups.
 | Table IIIMain results of the pooled data in
the meta-analysis. |
Table III
Main results of the pooled data in
the meta-analysis.
| Variable | No.
(cases/controls) | AA vs. GG
| (AA+AG) vs. GG
| AA vs. (AG+GG)
|
|---|
| OR (95% CI) | P-value | P-value
(Q-test) | OR (95% CI) | P-value | P-value
(Q-test) | OR (95% CI) | P-value | P-value
(Q-test) |
|---|
| Total | 1128/1276 | 1.06
(0.62–1.82) | 0.83 | 0.002 | 1.04
(0.76–1.43) | 0.80 | 0.08 | 1.06
(0.70–1.59) | 0.80 | 0.006 |
| Ethnicity | | | | | | | | | | |
| Caucasian | 212/346 | 0.84
(0.11–6.54) | 0.87 | 0.001 | 0.95
(0.31–2.90) | 0.93 | 0.02 | 0.84
(0.19–3.73) | 0.82 | 0.004 |
| Asian | 836/850 | 1.37
(0.97–1.95) | 0.08 | 0.3 | 1.18
(0.92–1.53) | 0.20 | 0.62 | 1.25
(0.88–1.79) | 0.22 | 0.16 |
| Mixed | 80/80 | 1.22
(0.56–2.66) | 0.62 | NA | 1.18
(0.61–2.29) | 0.61 | NA | 1.13
(0.57–2.22) | 0.73 | NA |
| Smoking status | | | | | | | | | | |
| Ever smoking | 602/588 | 0.55
(0.04–7.12) | 0.65 | 0.000 | 0.83
(0.32–2.20) | 0.71 | 0.002 | 0.62
(0.07–5.38) | 0.66 | 0.000 |
| Never
smoking | 188/342 | 1.16
(0.65–2.08) | 0.61 | 0.71 | 1.01
(0.61–1.67) | 0.96 | 0.75 | 1.22
(0.81–1.85) | 0.34 | 0.82 |
| Source of
control | | | | | | | | | | |
|
Hospital-based | 978/1103 | 1.09
(0.50–2.38) | 0.83 | 0.001 | 1.06
(0.68–1.66) | 0.78 | 0.03 | 1.06
(0.59–1.91) | 0.85 | 0.002 |
|
Population-based | 150/173 | 1.02
(0.56–1.84) | 0.96 | 0.49 | 1.02
(0.61–1.68) | 0.95 | 0.48 | 1.02
(0.63–1.65) | 0.95 | 0.67 |
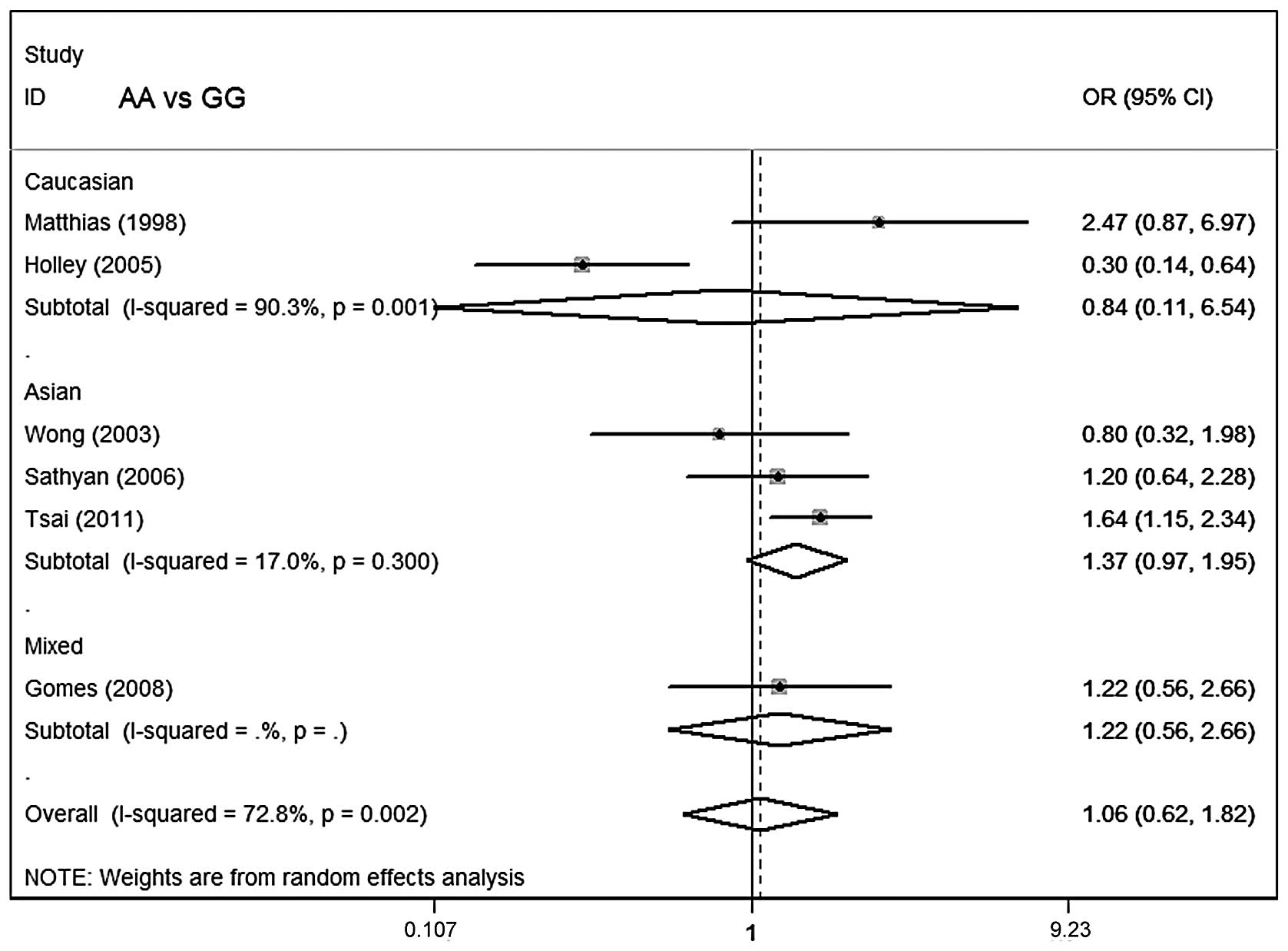
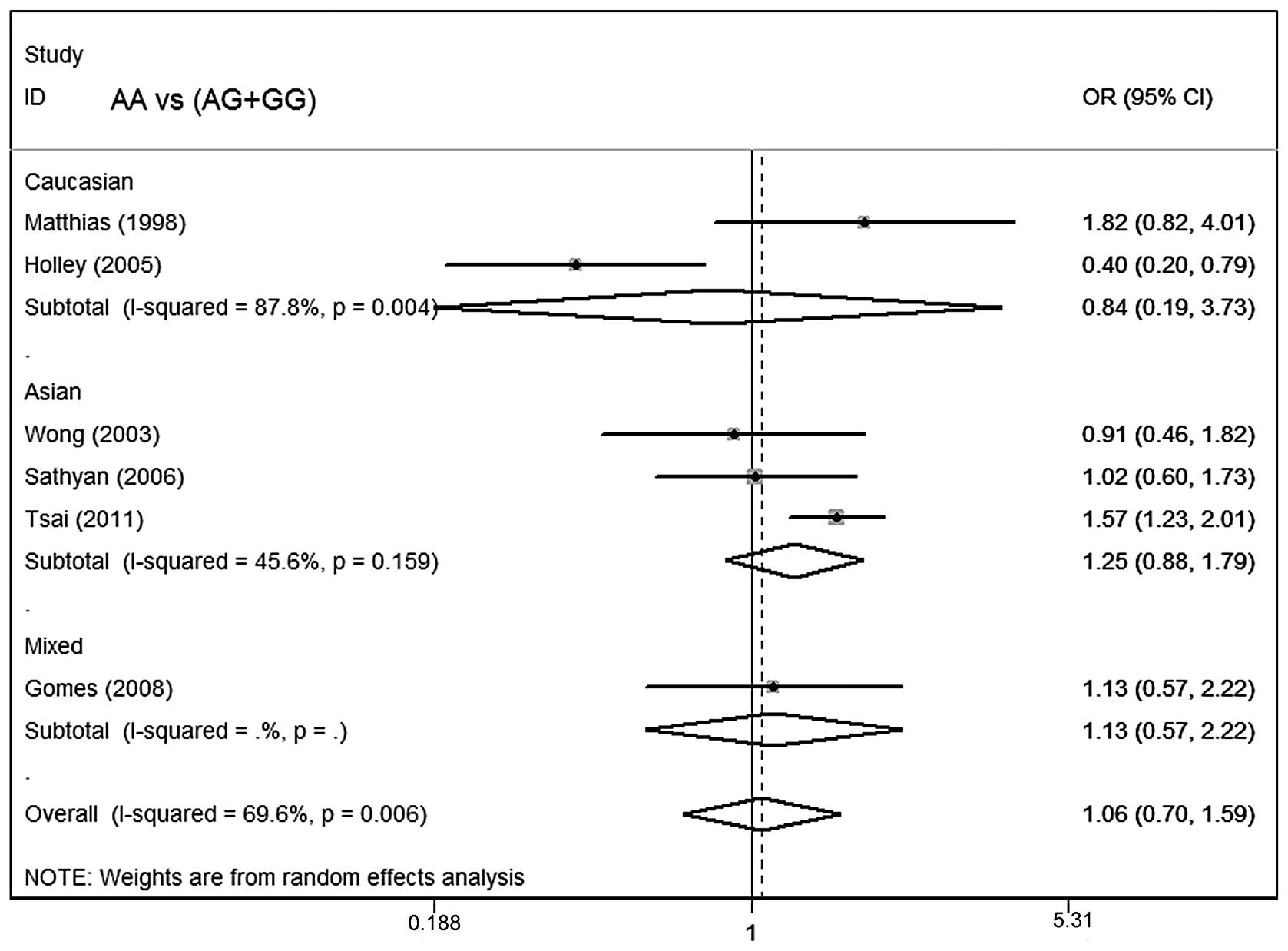
Meta-analysis results
Table III lists the
main results of the meta-analysis for oral cancer. The overall data
did not demonstrate any association of CCND1 polymorphism with oral
cancer risk (AA vs. GG: OR=1.06; 95% CI, 0.62–1.82; P=0.002 for
heterogeneity; dominant model: OR=1.04; 95% CI, 0.76–1.43; P=0.08
for heterogeneity; recessive model: OR=1.06; 95% CI, 0.70–1.59;
P=0.006 for heterogeneity; Figs.
1–3).
In subgroup analysis stratified by ethnicity, no
significant associations were identified among Asians, Caucasians
or mixed ethnicities for the three genetic models. In subgroup
analysis with regard to smoking status and control source, no
significant association of the CCND1 polymorphism with oral cancer
susceptibility was observed.
Sensitivity analysis
When the effects-models were changed, the
significance of the overall data for the three models was not
statistically altered (data not shown). We then excluded two
studies (29,30) whose genetic distributions in
controls exhibited a marked deviation from HWE, given that the
deviation may contribute to any bias (35). The significance of the overall data
in the three models was also not statistically altered, confirming
the stability of the results. Hence, results of the sensitivity
analysis suggest that the data in this meta-analysis are relatively
stable and credible.
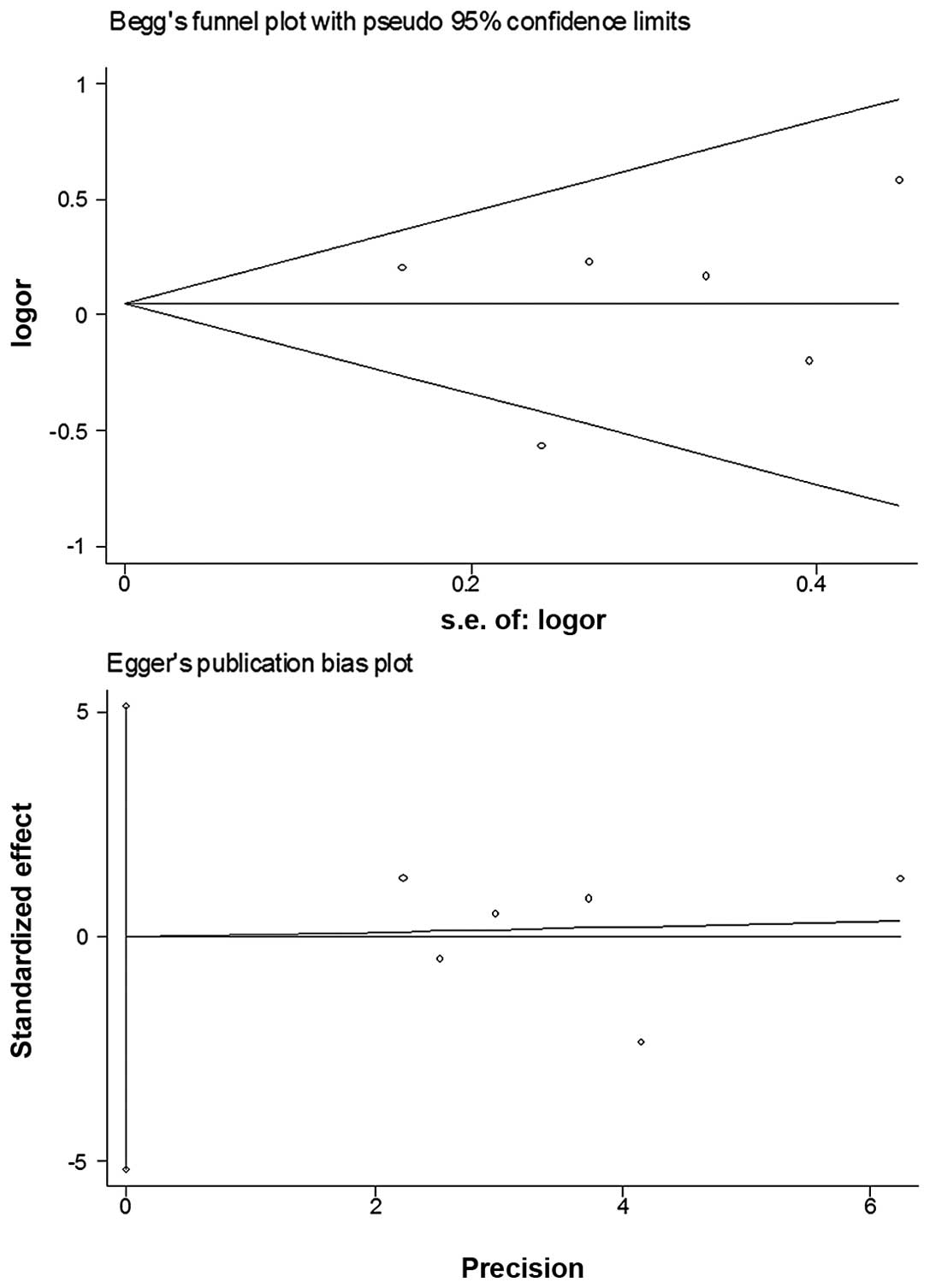
Bias diagnostics
Funnel plots were created for the assessment of
possible publication biases. Egger’s linear regression tests were
used to assess the symmetry of the plots (Fig. 4). The data suggest that the funnel
plots were symmetrical for the three models (AA vs. GG: t=−0.88,
P>0.05; dominant model: t=−0.01, P>0.05; recessive model:
t=−1.55, P>0.05), suggesting that the results of these
meta-analyses are relatively stable and the publication biases do
not have an evident influence on the results of the
meta-analyses.
Discussion
In the present study, the results of meta-analyses
failed to reveal a significant association of CCND1 G870A
polymorphism with oral cancer risk.
Recently, several meta-analyses have been conducted
to assess the association of CCND1 G870A polymorphisms with cancer
risks. The A allele has been suggested to increase risk of
digestive tract (36) and
colorectal cancer (37) in
Caucasian populations. Similarly, the A allele appears to increase
cancer risk in Chinese populations for breast (38) and bladder cancer (39), indicating that A allele carriers
may produce an elevated cancer risk. However, a meta-analysis
conducted on cervical cancer (40)
failed to show this marked association. Similarly, no evidence
supports the association of CCND1 genetic variation with head and
neck cancer (41) in a recently
published meta-analysis, of which the data were combined as head
and neck cancer rather than oral carcinoma, and two important
studies (29,32) on oral cancer that met the inclusion
criteria were ignored. In the present study, we focused on oral
cancer and screened the literature to include possible studies. In
the present study, in addition to ethnicity, more subgroups with
regard to smoking status and source of controls were further
assessed. No marked increased oral cancer risk was observed in any
comparisons for the three genetic models.
Smoking is a well-known risk factor for a number of
cancers, including lung and oral cancer. Effects of interactions
between smoking and the CCND1 genotype on lung cancer risk have
been suggested previously (42,43).
In this study, no increased cancer risk was demonstrated in the
smoking subgroup analysis (Table
III), suggesting that smoking did not modify the effect of
CCND1 polymorphisms on oral cancer risk. Notably, only two studies
(29,32) containing 790 cases and 930 controls
were available for the subgroup analysis. Further studies with
large sample sizes assessing smoking status are required for
further clarification.
Between-study heterogeneity was observed in Table III and thus random-effects models
were used. We further deselected the studies (29,30)
whose controls deviated from the HWE and observed that the
heterogeneity was not removed. Additionally, the significance of
the overall data was not statistically altered. Therefore, these
two studies were not excluded from the present study. However, the
P-value for heterogeneity indicated a reduced or removed
heterogeneity when the data were divided for subgroup analysis.
Publication biases were assessed via funnel plots
and their symmetries were further evaluated by Egger’s linear
regression tests. No evident biases were observed, suggesting
little influences of publication biases on the results.
Several limitations may be included in the present
study. First, in this meta-analysis, the majority of published
studies were written in English, Chinese and Portuguese, as cited
by the databases consulted. It is possible that certain published
or unpublished studies written in other languages that met the
inclusion criteria were missed. Hence, inevitable publication
biases may exist, although the funnel plots and Egger’s linear
regression tests implied no marked publication biases. Second, the
subgroup analysis concerned only Caucasians, Asians and mixed
ethnicities. Data with regard to other ethnicities were not
identified. The Asian populations were confined to Indians and
Chinese in Taiwan. Studies conducted in Southeast Asia, Japan and
mainland China are required to increase statistical power for the
demonstration of this association. Third, hospital-based controls
were used in some included studies. Hence, non-differential
misclassification bias may exist. However, subgroup analysis was
carried out and no evident influence on the results was identified.
Gene-gene and gene-environment interactions should also be
considered in further studies. However, the sensitivity analysis
and publication bias analysis showed the stability and credibility
of the present meta-analysis.
In summary, despite the limitations, the results of
the present meta-analysis failed to suggest a significant
association between CCND1 G870A genetic variations and oral cancer
risk.
Acknowledgements
This study was supported by the
Initial Fund for Doctors of Guiyang Medical College (2009–14) and
the China Postdoctoral Science Foundation (20100471772).
References
|
1
|
Adeyemi BF, Olusanya AA and Lawoyin JO:
Oral squamous cell carcinoma, socioeconomic status and history of
exposure to alcohol and tobacco. J Natl Med Assoc. 103:498–502.
2011.PubMed/NCBI
|
|
2
|
Li L, Psoter WJ, Buxó CJ, Elias A,
Cuadrado L and Morse DE: Smoking and drinking in relation to oral
potentially malignant disorders in Puerto Rico: a case-control
study. BMC Cancer. 11:3242011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin WJ, Jiang RS, Wu SH, Chen FJ and Liu
SA: Smoking, alcohol, and betel quid and oral cancer: a prospective
cohort study. J Oncol. 2011:5259762011.PubMed/NCBI
|
|
4
|
Zhuo W, Wang Y, Zhuo X, et al: CYP1A1 and
GSTM1 polymorphisms and oral cancer risk: association studies via
evidence-based meta-analyses. Cancer Invest. 27:86–95. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhuo XL, Li Q, Zhou Y, et al: Study on
TP53 codon 72 polymorphisms with oral carcinoma susceptibility.
Arch Med Res. 40:625–634. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou C, Zhou Y, Li J, et al: The Arg194Trp
polymorphism in the X-ray repair cross-complementing group 1 gene
as a potential risk factor of oral cancer: a meta-analysis. Tohoku
J Exp Med. 219:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niu Y, Hu Y, Wu M, et al: CYP2E1 Rsa I/Pst
I polymorphism contributes to oral cancer susceptibility: a
meta-analysis. Mol Biol Rep. 39:607–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Witzel II, Koh LF and Perkins ND:
Regulation of cyclin D1 gene expression. Biochem Soc Trans.
38:217–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reis-Filho JS, Savage K, Lambros MB, et
al: Cyclin D1 protein overexpression and CCND1 amplification in
breast carcinomas: an immunohistochemical and chromogenic in
situ hybridisation analysis. Mod Pathol. 19:999–1009. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar H, Vandana R and Kumar G:
Immunohistochemical expression of cyclin D1 in ameloblastomas and
adenomatoid odontogenic tumors. J Oral Maxillofac Pathol.
15:283–287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaminagakura E, Werneck da Cunha I, Soares
FA, Nishimoto IN and Kowalski LP: CCND1 amplification and protein
overexpression in oral squamous cell carcinoma of young patients.
Head Neck. 33:1413–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saini SS and Klein MA: Targeting Cyclin D1
in non-small cell lung cancer and mesothelioma cells by antisense
oligonucleotides. Anticancer Res. 31:3683–3690. 2011.PubMed/NCBI
|
|
13
|
Musgrove EA, Caldon CE, Barraclough J,
Stone A and Sutherland RL: Cyclin D as a therapeutic target in
cancer. Nat Rev Cancer. 11:558–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pabalan N, Bapat B, Sung L, Jarjanazi H,
Francisco-Pabalan O and Ozcelik H: Cyclin D1 Pro241Pro
(CCND1-G870A) polymorphism is associated with increased cancer risk
in human populations: a meta-analysis. Cancer Epidemiol Biomarkers
Prev. 17:2773–2781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Betticher DC, Thatcher N, Altermatt HJ,
Hoban P, Ryder WD and Heighway J: Alternate splicing produces a
novel cyclin D1 transcript. Oncogene. 11:1005–1011. 1995.PubMed/NCBI
|
|
16
|
Solomon DA, Wang Y, Fox SR, et al: Cyclin
D1 splice variants. Differential effects on localization, RB
phosphorylation, and cellular transformation. J Biol Chem.
278:30339–30347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sathyan KM, Nalinakumari KR, Abraham T and
Kannan S: CCND1 polymorphisms (A870G and C1722G) modulate its
protein expression and survival in oral carcinoma. Oral Oncol.
44:689–697. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rydzanicz M, Golusinski P,
Mielcarek-Kuchta D, Golusinski W and Szyfter K: Cyclin D1 gene
(CCND1) polymorphism and the risk of squamous cell carcinoma of the
larynx. Eur Arch Otorhinolaryngol. 263:43–48. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Munafò MR, Clark TG and Flint J: Assessing
publication bias in genetic association studies: evidence from a
recent meta-analysis. Psychiatry Res. 129:39–44. 2004.PubMed/NCBI
|
|
20
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishimoto IN, Pinheiro NA, Rogatto SR, et
al: Cyclin D1 gene polymorphism as a risk factor for squamous cell
carcinoma of the upper aerodigestive system in non-alcoholics. Oral
Oncol. 40:604–610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Izzo JG, Papadimitrakopoulou VA, Liu DD,
et al: Cyclin D1 genotype, response to biochemoprevention, and
progression rate to upper aerodigestive tract cancer. J Natl Cancer
Inst. 95:198–205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taniyama Y, Takeuchi S and Kuroda Y:
Genetic polymorphisms and oral cancer. J UOEH. 32:221–236.
2010.PubMed/NCBI
|
|
24
|
Henriksson E, Baldetorp B, Borg A, et al:
p53 mutation and cyclin D1 amplification correlate with cisplatin
sensitivity in xenografted human squamous cell carcinomas from head
and neck. Acta Oncol. 45:300–305. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu HS, Lu HH, Lui MT, et al: Detection of
copy number amplification of cyclin D1 (CCND1) and cortactin (CTTN)
in oral carcinoma and oral brushed samples from areca chewers. Oral
Oncol. 45:1032–1036. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuropkat C, Venkatesan TK, Caldarelli DD,
et al: Abnormalities of molecular regulators of proliferation and
apoptosis in carcinoma of the oral cavity and oropharynx. Auris
Nasus Larynx. 29:165–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sartor M, Steingrimsdottir H, Elamin F, et
al: Role of p16/MTS1, cyclin D1 and RB in primary oral cancer and
oral cancer cell lines. Br J Cancer. 80:79–86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu J, Gimenez-Conti IB, Cunningham JE, et
al: Alterations of p53, cyclin D1, Rb, and H-ras in human oral
carcinomas related to tobacco use. Cancer. 83:204–212. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsai MH, Tsai CW, Tsou YA, Hua CH, Hsu CF
and Bau DT: Significant association of cyclin D1 single nucleotide
polymorphisms with oral cancer in Taiwan. Anticancer Res.
31:227–231. 2011.PubMed/NCBI
|
|
30
|
Gomes CC, Drummond SN, Guimarães AL,
Andrade CI, Mesquita RA and Gomez RS: P21/WAF1 and cyclin D1
variants and oral squamous cell carcinoma. J Oral Pathol Med.
37:151–156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sathyan KM, Nalinakumari KR, Abraham T and
Kannan S: Influence of single nucleotide polymorphisms in H-Ras and
cyclin D1 genes on oral cancer susceptibility. Oral Oncol.
42:607–613. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holley SL, Matthias C, Jahnke V, Fryer AA,
Strange RC and Hoban PR: Association of cyclin D1 polymorphism with
increased susceptibility to oral squamous cell carcinoma. Oral
Oncol. 41:156–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong YK, Lin SC, Chang CS, et al: Cyclin
D1 genotype in areca-associated oral squamous cell carcinoma. J
Oral Pathol Med. 32:265–270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matthias C, Branigan K, Jahnke V, et al:
Polymorphism within the cyclin D1 gene is associated with prognosis
in patients with squamous cell carcinoma of the head and neck. Clin
Cancer Res. 4:2411–2418. 1998.PubMed/NCBI
|
|
35
|
Thakkinstian A, McElduff P, D’Este C,
Duffy D and Attia J: A method for meta-analysis of molecular
association studies. Stat Med. 24:1291–1306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen C, Sun H, Sun D, et al: Polymorphisms
of tumor necrosis factor-alpha and breast cancer risk: a
meta-analysis. Breast Cancer Res Treat. 126:763–770. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang LQ, Wang J, Shang JQ, et al: Cyclin
D1 G870A polymorphism and colorectal cancer susceptibility: a
meta-analysis of 20 populations. Int J Colorectal Dis.
26:1249–1255. 2011. View Article : Google Scholar
|
|
38
|
Sergentanis TN and Economopoulos KP:
Cyclin D1 G870A polymorphism and breast cancer risk: a
meta-analysis comprising 9,911 cases and 11,171 controls. Mol Biol
Rep. 38:4955–4963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yuan L, Gu X, Shao J, Wang M, Zhu Q and
Zhang Z: Cyclin D1 G870A polymorphism is associated with risk and
clinicopathologic characteristics of bladder cancer. DNA Cell Biol.
29:611–617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ni J, Wang M, Wang M, Fu S, Zhou D, Zhang
Z and Han S: CCND1 G870A polymorphism and cervical cancer risk: a
case-control study and meta-analysis. J Cancer Res Clin Oncol.
137:489–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang C, Wang Z, Yu J, Wu Y, Zhu Z and Chen
N: CCND1 G870A polymorphism and risk for head and neck cancer: a
meta-analysis. Med Oncol. 28:1319–1324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsia TC, Liu CJ, Lin CH, et al:
Interaction of CCND1 genotype and smoking habit in Taiwan lung
cancer patients. Anticancer Res. 31:3601–3605. 2011.PubMed/NCBI
|
|
43
|
Gautschi O, Hugli B, Ziegler A, et al:
Cyclin D1 (CCND1) A870G gene polymorphism modulates smoking-induced
lung cancer risk and response to platinum-based chemotherapy in
non-small cell lung cancer (NSCLC) patients. Lung Cancer.
51:303–311. 2006. View Article : Google Scholar : PubMed/NCBI
|