Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies worldwide, particularly in developing
countries, including China (1,2). Due
to its aggressive nature and the lack of means for early diagnosis
and effective therapy, the HCC mortality rate remains high. Various
novel therapeutic approaches are under extensive investigation,
among which targeted gene therapy is a potential candidate with
promising therapeutic effect.
Introducing a specific tumor suppressor or a gene
with tumor-suppressive function, including inhibition of growth,
invasion/metastasis and inducing apoptosis, is a routine method of
cancer gene therapy. Blocking overexpressed oncogenic genes is
another method for cancer gene therapy. Antisense technology has
been developed, which demonstrates potential for this purpose.
Since the discovery of RNA interference (RNAi) (3,4),
small interfering RNA (siRNA)-based technology is gradually
replacing antisense technology due to its more potent and specific
effect in silencing target gene expression (5,6). As
a newly developed method, the use of artificial microRNAs
(amiRNAs), also known as the second generation of short hairpin RNA
(shRNA), has been shown to be more convenient, efficient and safe
by a number of investigators compared with chemically synthesized
siRNA or shRNA (7,8). To maximize the potential of amiRNA in
cancer gene therapy, the mechanism of the expression and processing
of amiRNA precursors has been studied in detail (9,10).
The amount of a specific mature microRNA (miRNA) in
a cell is regulated at transcriptional and post-transcriptional
levels. For gene therapy of HCC, the transcription of a therapeutic
amiRNA precursor is usually controlled by the cancer-specific
α-fetoprotein promoter (AFP) for targeted expression (11,12),
while the post-transcriptional regulation of the amiRNA depends not
only on cellular processing machinery, but also on the specific
flanking sequence surrounding the cleavage sites, which varies
significantly (9,10,13).
The specific cellular processing machinery in a HCC cell and the
specific sequence of the miRNA precursor determine the quantity of
the specific mature miRNA. Therefore we postulate that the sequence
of highly abundant miRNAs in HCC cells would be favorable
processing targets and should be explored for their potential in
potent HCC-specific amiRNA construction. In the present study, we
evaluated the processing efficiencies of the precursors of six
natural miRNAs with high abundances in HCC cells, including
miR-18a, miR-21, miR-192, miR-221, miR-222 and miR-224 (14–19),
by constructing amiRNAs targeting firefly luciferase (20). These were then cotransfected with a
luciferase expression vector into human HCC cells, Hep3B and HepG2.
The most efficient miR-221 precursor sequence was determined.
Materials and methods
Cell lines
Human HCC cell lines Hep3B and HepG2 were purchased
from American Type Culture Collection (ATCC, Manassas, VA, USA),
supplemented with 10% bovine growth serum (Thermo Scientific, Inc.,
Waltham, MA, USA) at 37°C and 5% CO2 under saturated
humidity.
Design and cloning of pre-amiRNAs
Precursors of six natural miRNAs with high abundance
in HCC cells were selected according to the literature, including
miR-18a (miRBase accession number: MI0000072), miR-21 (miRBase
accession number: MI0000077), miR-192 (miRBase accession number:
MI0000234), miR-221 (miRBase accession number: MI0000298), miR-222
(miRBase accession number: MI0000299) and miR-224 (miRBase
accession number: MI0000301). The sequence specifically targeting
the firefly luciferase gene (luc: 5′-cgc ctg aag tct ctg att aa-3′)
(20) was introduced into the
precursors to substitute each of the core sequences (Fig. 1). All artificial pre-miRNAs were
cloned by polymerase chain reaction (PCR) with each of the primers
(Table I). Pre-miR-18a-luc and
pre-miR-21-luc were obtained by one round of PCR. Pre-miR-192-luc,
pre-miR-221-luc, pre-miR-222-luc and pre-miR-224-luc were obtained
by two rounds of PCR using F1 and R1 as primers for the first round
PCR. For the second round of PCR, pre-miR-224-luc used F2 and R1 as
primers, while pre-miR-192-luc, pre-miR-221-luc and pre-miR-222-luc
used F2 and R2. The PCR conditions are listed in Table II. PCR products were separated by
3% agarose gel electrophoresis and recovered using a QIAquick Gel
Extraction kit (Qiagen, Hilden, Germany) and cloned into a pMD19-T
vector (Takara Bio Inc., Shiga, Japan). The sequences were verified
by DNA sequencing and subcloned into the mammalian expression
vector pIRES2-EGFP (Clontech Laboratories Inc., Mountain View, CA,
USA) at the sites BglII and SacII to generate various
amiRNA precursor-expressing vectors.
 | Table I.Primers for amplifying amiRNA
precursors. |
Table I.
Primers for amplifying amiRNA
precursors.
| Pre-amiRNA | Primer sequence |
|---|
| Pre-miR-18a-luc | F:
5′-agatctgatcctgttcttaatcagagacttcaggcggagtgaagtagattagcatctcg-3′ |
| R:
5′-ccgcggatcgtagtgccagtaatcagagacttcaggcgagatgctaatctacttcact-3′ |
| Pre-miR-21-luc | F:
5′-agatctgatcctgtcgggttaatcagagacttcaggcggactgttgaatctcatggcc-3′ |
| R:
5′-ccgcggatcgtagtgtcagactaatcagagacttcaggcggccatgagattcaacagtc-3′ |
| Pre-miR-192-luc | F1:
5′-accgagtgcacagggctttaatcagagacttcaggcccagtgctctcgtctcccctctg-3′ |
| R1:
5′-cattgaggcgaacatacctgtaatcagagacttcaggcccagaggggagacgagagcac-3′ |
| F2:
5′-agatctgatccgccgagaccgagtgcacagggcttta 3′ |
| R2:
5′-ccgcggatcgtaggctggcattgaggcgaacatacctg 3′ |
| Pre-miR-221-luc | F1
5′-aggtctggggcatgaccgcctgaagtctctgattatttaagtgttcgttaggcaactta-3′ |
| R1:
5′-tgtttccaggtagcctgaccgcctgaagtctctgattaagttgcctaacgaacacttaa-3′ |
| F2
5′-agatctgatcctgaacatccaggtctggggcatgaccgcc-3′ |
| R2
5′-ccgcggatcgtaggagaacatgtttccaggtagcctgacc-3′ |
| Pre-miR-222-luc | F1
5′-aggtgtaggtaccctcaatggcgcctgaagtctctgattatcctgtctttcgtaatcag-3′ |
| R1
5′-aagatgccatcagagacgcctgaagtctctgattaagctgattacgaaagacaggataa-3′ |
| F2
5′-agatctgatccgctgctggaaggtgtaggtaccctcaatg-3′ |
| R2
5′-ccgcggatcgtagagctagaagatgccatcagagacgcc-3′ |
|
Pre-miR-224-luc | F1:
5′-gggctttttaatcagagacttcaggcggtagtagatgattgtgcattgtttcaaccgcc-3′ |
| R1:
5′-ccgcggatcgtaggggctttggaatcagagacttcaggcggttgaaacaatgcacaatc-3′ |
| F2:
5′-agatctgatccgggctttttaatcagagacttc-3′ |
 | Table II.PCR parameters for amiRNA
precursors. |
Table II.
PCR parameters for amiRNA
precursors.
| Pre-amiRNA | PCR parameters | Product size
(bp) |
|---|
|
Pre-miR-18a-luc | 95°C, 2 min; 95°C,
30 sec, 57°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5min | 95 |
| Pre-miR-21-luc | 95°C, 2 min; 95°C,
30 sec, 55°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5min | 97 |
|
Pre-miR-192-luc | (1st ) 95°C, 2 min;
95°C, 30 sec, 61°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 98 |
| (2nd) 95°C, 2 min;
95°C, 30 sec, 55°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 133 |
|
Pre-miR-221-luc | (1st ) 95°C, 2 min;
95°C, 30 sec, 55°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 95 |
| (2nd) 95°C, 2 min;
95°C, 30 sec, 57°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 135 |
|
Pre-miR-222-luc | (1st ) 95°C, 2 min;
95°C, 30 sec, 55°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 96 |
| (2nd) 95°C, 2 min;
95°C, 30 sec, 57°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 135 |
|
Pre-miR-224-luc | (1st ) 95°C, 2 min;
95°C, 30 sec, 63°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 95 |
| (2nd) 95°C, 2 min;
95°C, 30 sec, 57°C, 30 sec, 72°C, 30 sec, 30 cycles; 72°C, 5
min | 106 |
Construction of the luciferase expression
vector
The human thymidine kinase (TK) promoter was cloned
by PCR with primer pairs 5′-ctc gag aaa tga gtc ttc gga cct cgc-3′
(forward) and 5′-aga tct tta agc ggg tcg ctg cag g-3′ (reverse)
using the plasmid pGL4.74[hRluc/TK] (Promega Corporation,
Madison, WI, USA) as the template and the following cycling
conditions: pre-denaturing at 95°C for 2 min followed by 30 cycles
of 95°C for 30 sec, 57°C for 30 sec and 72°C for 45 sec, and a
final extension at 72°C for 5 min. The 765 bp PCR product was
cloned into the pMD19-T vector (Takara Bio Inc.) and subcloned into
the luciferase reporter vector pGL3.0-basic at sites BglII
and XholI. The sequence was verified by DNA sequencing.
Co-transfection of the luciferase
reporter vector and amiRNA precursor-expressing vectors
HCC cells were seeded into 24-well plates with
3×105 cells/well or 6-well plates with 9×105
cells/well. The amiRNA precursor-expressing vectors or empty
control vector (pIRES2-EGFP plasmids) and a typical 100 μl
transfection mixture was prepared with 1.5 μg plasmid DNA
(pIRES2-EGFP: pGL3.0-basic/TK: pGL4.74[hRluc/TK] in a
1:5:0.05 ratio) and 3 μl transfection reagent Lipofectamine
LTX (Invitrogen Life Technologies, Carlsbad, CA, USA) according to
the manufacturer’s instructions. The transfection mixture was added
to cultured cells in triplicate with 100 μl/well for 24-well
plates or 300 μl/well for 6-well plates. The transfection
mixture-containing medium was replaced by fresh medium after 24
h.
Knockdown efficacy of amiRNA by
dual-luciferase assay
A luciferase assay was performed 48 h after
transfection to evaluate the efficacy of each of the amiRNAs at the
protein activity level using a Dual-Luciferase® Reporter
1000 Assay kit (Promega) and GloMax® 20/20 Luminometer
(Promega) according to the manufacturer’s instructions. Briefly,
transfected cells in 24-well plates were washed with ice-cold
phosphate-buffered saline (PBS), lysed in 100 μl 1X passive
lysis buffer (PLB) and the lysate was collected. Then, 100
μl luciferase assay reagent II was added to 20 μl
lysate and mixed rapidly, followed by 10-sec measurements for
firefly luciferase activity. Then, 100 μl Stop and
Glo® reagent was added and mixed rapidly, followed by
10-sec measurements for Renilla luciferase activity. The
relative luciferase unit (RLU) was calculated as the ratio of
firefly luciferase activity to Renilla luciferase activity.
Relative RLU (RLUsample/RLUblank) was used to
evaluate the knockdown efficacy.
Reverse transcription (RT)-PCR
RT-PCR was performed 48 h after transfection to
evaluate the knockdown efficacy of amiRNAs at the mRNA level. Total
RNA of transfected cells in 6-well plates was extracted using
TRIzol reagent (Invitrogen Life Technologies) and treated with RQ1
RNase-Free DNase (Promega) to remove any contamination of DNA. For
each sample, 2 μg total RNA was used for RT with M-MLV
Reverse Transcriptase (Promega). The primers for PCR are listed in
Table III and the following
cycling parameters were used: 95°C for 2 min; 95°C for 30 sec, 60°C
for 30 sec, 72°C for 30 sec for 26 cycles (for firefly luciferase,
product size 200 bp) and 35 cycles (for Renilla luciferase,
product size 150 bp). PCR products were examined by 2% agarose gel
electrophoresis and images were documented using
FluorChem® FC2 Imager (Alpha Innotech, San Leandro, CA,
USA). The integrated volume of each band was quantified by
AlphaView SA software (Alpha Innotech). The relative expression of
firefly luciferase mRNA (ReLuc) was calculated as the integrated
volume ratio of the PCR product band of firefly luciferase to that
of Renilla luciferase normalized with the blank control
sample.
 | Table III.Primers for reverse
transcription-polymerase chain reaction (RT-PCR) of firefly and
Renilla luciferase. |
Table III.
Primers for reverse
transcription-polymerase chain reaction (RT-PCR) of firefly and
Renilla luciferase.
| Target | Primer
sequence |
|---|
| Firefly
luciferase | F:
5′-cgccgccgttgttgttttgga-3′ |
| R:
5′-tctttccgcccttcttggcct-3′ |
| Renilla
luciferase | F:
5′-agtccgaccctgggttcttttcca-3′ |
| R:
5′-cgcgctccacgaagctcttgat-3′ |
Real-time quantitative PCR
The relative expression levels of pre-amiRNAs and
mature amiRNAs were determined by real-time quantitative PCR using
a miScript RT kit (Qiagen) and miScript SYBR®-Green PCR
kit (Qiagen) on an ABI 7500 Fast instrument (Applied Biosystems,
Foster City, CA, USA), with the Renilla luciferase mRNA
level as the internal normalization control. The relative
expression level was evaluated with the ΔCt method (ΔCt =
Ctpre-amiRNA or mature amiRNA -
CtRenillaluciferase). The primers and
cycling parameters used are listed in Table IV.
 | Table IV.Primers and parameters for real-time
quantitative polymerase chain reaction (PCR). |
Table IV.
Primers and parameters for real-time
quantitative polymerase chain reaction (PCR).
| Target | Primer
sequence | Cycling
parameters |
|---|
|
Pre-miR-221-luc | F:
5′-ctctgattatttaagtgttcg-3′ | 95°C, 15 min |
| 94°C, 15 sec; 55°C,
30 sec; 70°C, 34 sec; 45 cycles |
| miR-221-luc | F:
5′-ttaatcagagacttcagg-3′ | 95°C, 15 min |
| 94°C, 15 sec; 55°C,
30 sec; 70°C, 34 sec; 45 cycles |
| Renilla
luciferase | F:
5′-agtccgaccctgggttcttttcca-3′ | 95°C, 15 min |
| R:
5′-cgcgctccacgaagctcttgat-3′ | 94°C, 15 sec; 60°C,
30 sec; 72°C, 34 sec; 45 cycles |
Results
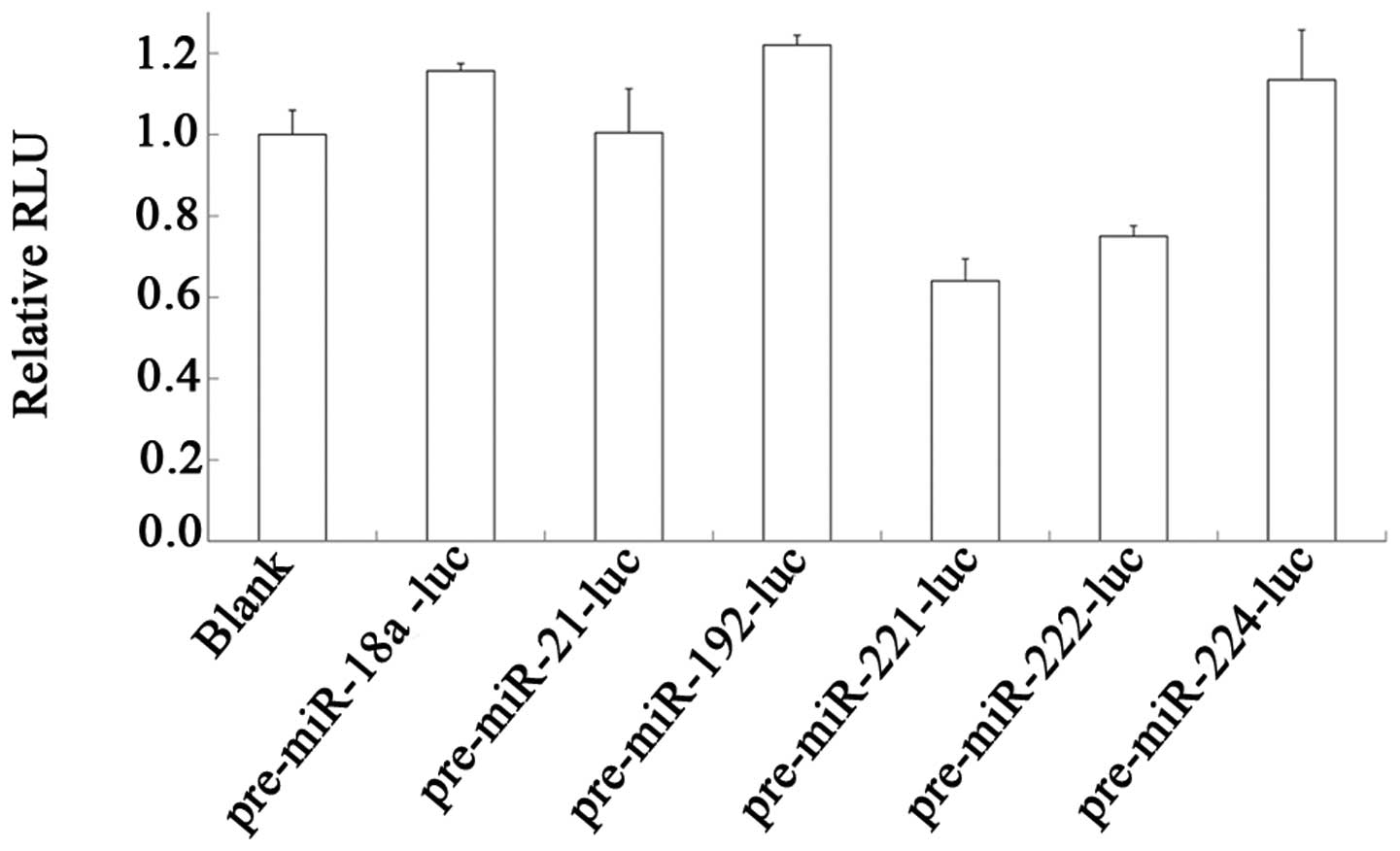
Knockdown efficiency of amiRNAs with
different precusor backbones in Hep3B cells
A total of six amiRNA precursors targeting the
firefly luciferase gene with different backbones were successfully
cloned and used for construction of an expression vector with
pIRES2-EGFP. The firefly luciferase expression vector was
constructed by placing the promoter of the human TK gene before the
luciferase gene in the pGL3.0-basic vector. The mRNAs of the amiRNA
precursor and firefly luciferase were expressed when co-transfected
into the HCC cells and mRNA of firefly luciferase was silenced by
the processed mature amiRNA. We analyzed the knockdown efficiencies
of the six amiRNA precursors by the co-transfection of the vectors
with the transfection control vector pGL4.74[hRluc/TK] in
Hep3B cells. The dual luciferase assay demonstrated that not all
the amiRNA precursors were efficiently processed to generate
amiRNAs targeting firefly luciferase mRNA (Fig. 2). The amiRNA precursor with the
backbone of miR-221 (pre-miR-221-luc) demonstrated the most
efficient processing.
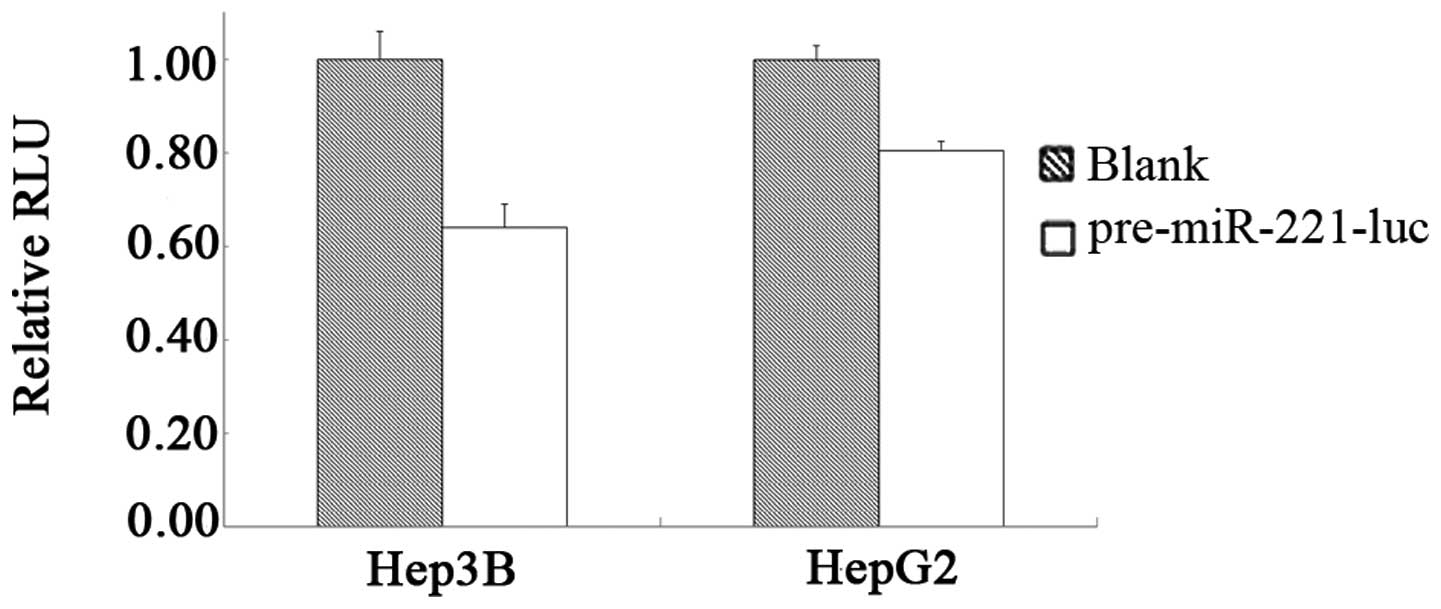
Knockdown efficiency of pre-miR-221-luc
in different cells
The processing and knockdown efficiency of
miR-221-luc in HCC Hep3B or HepG2 cells was examined by dual
luciferase assay. As shown in Fig.
3, pre-miR-221-luc was processed in HCC Hep3B and HepG2 cells
and the knockdown of luciferase was observed with RLUs of 64.04 and
80.48%, respectively, compared with the controls. This is a
relative knockdown analysis with co-transfected
luciferase-expressing vector; the knockdown efficiency may vary
when altering the amount of the co-transfected
luciferase-expressing vector and the transfection-control vector.
The knockdown efficiency of a specific endogenous target gene when
applying this precursor structure in gene therapy study was
assessed individually.
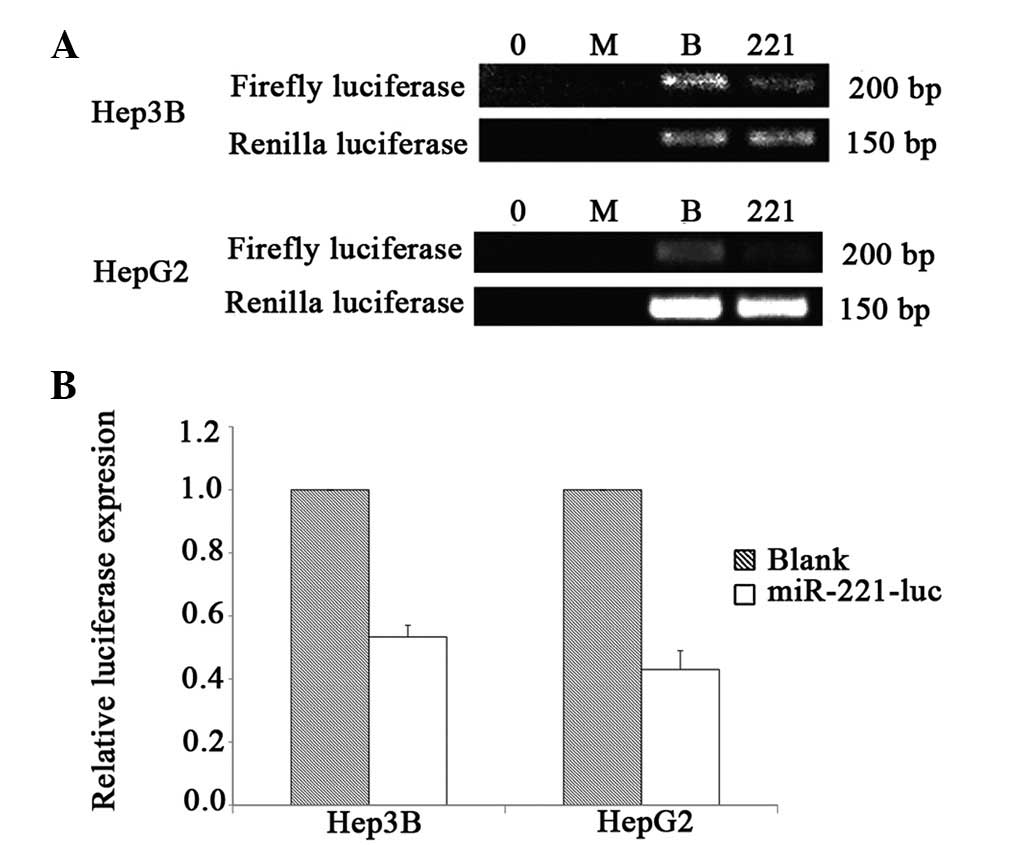
We further verified the knockdown efficiency by
RT-PCR using Renilla luciferase as the control. As shown in
Fig. 4, pre-miR-221-luc was
efficiently processed and led to 46.65% and 57.00% knockdown
efficiencies in Hep3B and HepG2 cells, respectively.
Expression and processing of miR-221-luc
by real-time quantitative PCR
To evaluate the expression and processing of
miR-221-luc in HCC cells, real-time quantitative PCR was used to
examine the level of amiRNA precursors and mature amiRNAs, with
respect to the normalization control, Renilla luciferase
mRNA. As shown in Table V, HCC
cells exhibited satisfactory expression of pre-miR-221-luc, shown
by ΔΔCt (−8.15 and −12.35 in Hep3B cells and HepG2 cells,
respectively), which represented the relative ratio of the level of
pre-miR-221-luc in HCC cells transfected with the
pre-miR-221-luc-expressing vector to that in non-transfected HCC
cells (the smaller the ΔΔCt value, the higher the pre-miR-221-luc
level). The expressed precursors were also processed efficiently to
mature miR-221-luc, shown by ΔΔCt (−7.32 and −12.17 in Hep3B cells
and HepG2 cells, respectively), which represents the relative ratio
of the level mature miR-221-luc in HCC cells transfected with the
pre-miR-221-luc-expressing vector to that in non-transfected HCC
cells (the smaller the ΔΔCt value, the higher the miR-221-luc
level).
 | Table V.Relative levels of pre-miR-221-luc
and mature miR-221-luc. |
Table V.
Relative levels of pre-miR-221-luc
and mature miR-221-luc.
| ΔCt
|
|---|
pre-miR-221-luc
| miR-221-luc
|
|---|
| mean | SD | mean | SD |
|---|
| Hep3B blank | 4.21 | 0.43 | 7.97 | 0.35 |
| Hep3B
pre-miR-221-luc | −3.94 | 0.57 | 0.65 | 0.70 |
| ΔΔCt | −8.15 | | −7.32 | |
| HepG2 blank | 17.24 | 0.54 | 17.93 | 0.77 |
| HepG2
pre-miR-221-luc | 4.89 | 0.47 | 5.76 | 0.42 |
| ΔΔCt | −12.35 | | −12.17 | |
Discussion
As the population increases and problems, including
ageing and environmental pollution by various carcinogens arise,
the incidence of cancer also presents a rapid increase; it ranks
first in developed countries and second in developing countries as
the leading cause of mortality (1). There were 12,700,000 new cases of
cancer in 2008 and 7,600,000 cancer mortalities. As one of the most
common types of cancer, the number of new cases of HCC is 748,300
and the number of mortalities due to HCC is 695,900 (2). Approximately half of the incidence
and mortalities of HCC occur in China (1). The management of HCC patients is
extremely costly in terms of medical resources. The therapeutic
effects of surgery, chemotherapy and radiotherapy and the prognosis
of HCC are not ideal. New means of therapy need to be explored.
Biotherapy, the so-called ‘fourth therapeutic model’
of malignancies, is showing its potential in clinics. Biotherapy
includes gene therapy, immunotherapy, anti-angiogenesis therapy,
oncolytic viral therapy and stem cell therapy (21–25).
Among these, gene therapy is one of the most important components
of biotherapy and is the main focus of research.
One of the approaches adopted in cancer gene therapy
is to block the overexpressed oncogenic genes in cancer cells. The
development of RNA interference (RNAi) technology provides an
effective method in this regard, which appears to be promising in
cancer gene therapy (26–28). Synthetic siRNAs and amiRNAs are
useful tools; amiRNA technology has been successfully used in gene
therapy studies. Compared with shRNA or siRNA, amiRNA has the
advantage of regulatable expression, more efficient processing
in vivo and greater safety as demonstrated by animal models
(7,8).
amiRNA is a technology that utilizes the framework
of precursors of natural miRNAs as the backbone with a specific
sequence targeting the gene of interest (29,30).
For naturally-occurring miRNAs, the quantity of a specific mature
miRNA is regulated at transcriptional and post-transcriptional
processing levels, and the processing efficiency depends on the
structure of its precursor (9,10).
The framework of the amiRNA precursor is critical for its
processing to obtain mature amiRNA, while the specific core
sequence to be processed to mature miRNA is critical for its
knockdown efficiency. Different frameworks may have different
processing efficiencies in different types of cell. The framework
of the precursor of miR-30 is widely used for its high efficiency
in processing in a number of cell types. For gene therapy, however,
the property of specifically targeted expression and processing of
an amiRNA is preferable to one with universal expression and
processing. Studies have demonstrated that an 11-base pair flanking
sequence of pre-miRNA stem structure is important for the
recognition and splicing by the Drosha-DGCR8 complex (13,31).
Impaired structure in this region may affect the quantity of mature
miRNA and the knockdown efficiency (31,32).
A number of studies on the clinical significance of
various miRNAs in HCC have been conducted (18,22,33);
however, there has been no report concerning the cause for the
difference of the levels of miRNAs.
In the present study, we selected six miRNAs that
were reported to be abundantly expressed in HCC cells and analyzed
the processing efficiency of the precursors of these miRNAs by
replacing the gene-specific sequence with a luciferase-targeting
sequence in the framework of the precursors. The expression of the
amiRNA precursor was controlled by the cytomegalovirus (CMV)
promoter to provide a high and uniform level of expression. The
dual luciferase reporter assay and RT-PCR reflected the consequence
of the expression and processing of amiRNA precursors on the target
firefly luciferase activity relative to the control Renilla
luciferase activity, and real-time quantitative PCR revealed the
levels of precursors and processed mature amiRNAs. Considering the
knockdown of an endogenous gene may have an unknown impact on the
cell, which may bring changes to numerous aspects, including
cellular machineries and the expression and/or processing of
amiRNA, we used firefly luciferase as the target gene for amiRNA in
this study. To minimize the effect of uneven transfection
efficiency on the expression of the firefly luciferase gene and
amiRNA precursors, we co-transfected a Renilla
luciferase-expression vector as a normalization standard.
Our results demonstrated that among all the amiRNA
precursors we analyzed, the one based on the miR-221 precursor
framework has the most potent knockdown effect on firefly
luciferase activity in HCC cells. Results from real-time
quantitative PCR revealed the expression and processing of amiRNA
in HCC cells, confirming that the amiRNA precursor based on the
miR-221 precursor is efficiently processed.
Acknowledgements
This study was supported by grants
from the Zhejiang Provincial Natural Science Foundation of China
(LZ12H16003) and the Foundation of Key Medical Sciences of Public
Health of Zhejiang Province (11-ZC02). The authors are very
grateful to Dr Jiang Cao of the Clinical Research Center, The
Second Affiliated Hospital, School of Medicine, Zhejiang University
for his helpful and critical comments on this manuscript. The
authors also thank Dr Jing Jia of the Center of Molecular Medicine,
Zhejiang Academy of Medical Sciences for her comments and
suggestions in the manuscript; Zhaoqiang Jiang of the Institute of
Hygiene, Zhejiang Academy of Medical Sciences for assistance in
statistical analysis; and Xiang Chen and Jiacong Zhao of the School
of Laboratory Medicine and Life Science, Wenzhou Medical University
for their technical assistance.
References
|
1.
|
Mathers C, Fat DM and Boerma J: The global
burden of disease: 2004 update. World Health Organization.
2008.
|
|
2.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Campbell TN and Choy FY: RNA interference:
past, present and future. Curr Issues Mol Biol. 7:1–6.
2005.PubMed/NCBI
|
|
4.
|
Sledz CA and Williams BRG: RNA
interference in biology and disease. Blood. 106:787–794. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Que XY, Li Y, Han Y and Li XZ: Effects of
siRNA-mediated Cdc2 silencing on MG63 cell proliferation and
apoptosis. Mol Med Rep. 7:466–470. 2013.PubMed/NCBI
|
|
6.
|
Yin Y, Chen X, Zhang CD, et al: Asymmetric
siRNA targeting the bcl-2 gene inhibits the proliferation of cancer
cells in vitro and in vivo. Int J Oncol. 42:253–260.
2013.PubMed/NCBI
|
|
7.
|
Boudreau RL, Martins I and Davidson BL:
Artificial microRNAs as siRNA shuttles: improved safety as compared
to shRNAs in vitro and in vivo. Mol Ther. 17:169–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liu X, Fang H, Chen H, et al: An
artificial miRNA against HPSE suppresses melanoma invasion
properties, correlating with a down-regulation of chemokines and
MAPK phosphorylation. PLoS One. 7:e386592012. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Slezak-Prochazka I, Durmus S, Kroesen BJ
and van den Berg A: MicroRNAs, macrocontrol: regulation of miRNA
processing. RNA. 16:1087–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Starega-Roslan J, Koscianska E, Kozlowski
P and Krzyzosiak WJ: The role of the precursor structure in the
biogenesis of microRNA. Cell Mol Life Sci. 68:2859–2871. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Zhang KJ, Qian J, Wang SB and Yang Y:
Targeting Gene-Viro-Therapy with AFP driving Apoptin gene shows
potent antitumor effect in hepatocarcinoma. J Biomed Sci.
19:202012. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Zhang KJ, Zhang J, Wu YM, et al: Complete
eradication of hepatomas using an oncolytic adenovirus containing
AFP promoter controlling E1A and an E1B deletion to drive IL-24
expression. Cancer Gene Ther. 19:619–629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Han J, Lee Y, Yeom KH, et al: Molecular
basis for the recognition of primary microRNAs by the Drosha-DGCR8
complex. Cell. 125:887–901. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Wang Y, Lee AT, Ma JZ, et al: Profiling
microRNA expression in hepatocellular carcinoma reveals
microRNA-224 up-regulation and apoptosis inhibitor-5 as a
microRNA-224-specific target. J Biol Chem. 283:13205–13215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Bala S, Marcos M and Szabo G: Emerging
role of microRNAs in liver diseases. World J Gastroenterol.
15:5633–5640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Boni V, Bandres E, Zarate R, Colucci G,
Maiello E and Garcia-Foncillas J: MicroRNAs as a new potential
therapeutic opportunity in gastrointestinal cancer. Oncology.
77(Suppl 1): 75–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Li M, Li J, Ding X, He M and Cheng SY:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar
|
|
18.
|
Ishida H, Tatsumi T, Hosui A, et al:
Alterations in microRNA expression profile in HCV-infected hepatoma
cells: involvement of miR-491 in regulation of HCV replication via
the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 412:92–97.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Albulescu R, Neagu M, Albulescu L and
Tanase C: Tissular and soluble miRNAs for diagnostic and therapy
improvement in digestive tract cancers. Expert Rev Mol Diagn.
11:101–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Silva JM, Li MZ, Chang K, et al:
Second-generation shRNA libraries covering the mouse and human
genomes. Nat Genet. 37:1281–1288. 2005.PubMed/NCBI
|
|
21.
|
Xu C, Lee SA and Chen X: RNA interference
as therapeutics for hepatocellular carcinoma. Recent Pat Anticancer
Drug Discov. 6:106–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Rutella S, Iudicone P, Bonanno G, et al:
Adoptive immunotherapy with cytokine-induced killer cells generated
with a new good manufacturing practice-grade protocol. Cytotherapy.
14:841–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Shojaei F: Anti-angiogenesis therapy in
cancer: current challenges and future perspectives. Cancer Lett.
320:130–137. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zeyaullah M, Patro M, Ahmad I, et al:
Oncolytic viruses in the treatment of cancer: a review of current
strategies. Pathol Oncol Res. 18:771–781. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Yao N, Yao D, Wang L, et al: Inhibition of
autocrine IGF-II on effect of human HepG2 cell proliferation and
angiogenesis factor expression. Tumour Biol. 33:1767–1776. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ibrahim AF, Weirauch U, Thomas M,
Grunweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Shi ZM, Wang J, Yan Z, et al: MiR-128
inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS
One. 7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Stegmeier F, Hu G, Rickles RJ, Hannon GJ
and Elledge SJ: A lentiviral microRNA-based system for single-copy
polymerase II-regulated RNA interference in mammalian cells. Proc
Natl Acad Sci USA. 102:13212–13217. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Chang K, Elledge SJ and Hannon GJ: Lessons
from Nature: microRNA-based shRNA libraries. Nat Methods.
3:707–714. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Hinton TM, Wise TG, Cottee PA and Doran
TJ: Native microRNA loop sequences can improve short hairpin RNA
processing for virus gene silencing in animal cells. J RNAi Gene
Silencing. 4:295–301. 2008.PubMed/NCBI
|
|
32.
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Augello C, Vaira V, Caruso L, et al:
MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster
as a novel prognostic biomarker in hepatocellular carcinoma. Liver
Int. 32:772–782. 2012. View Article : Google Scholar : PubMed/NCBI
|