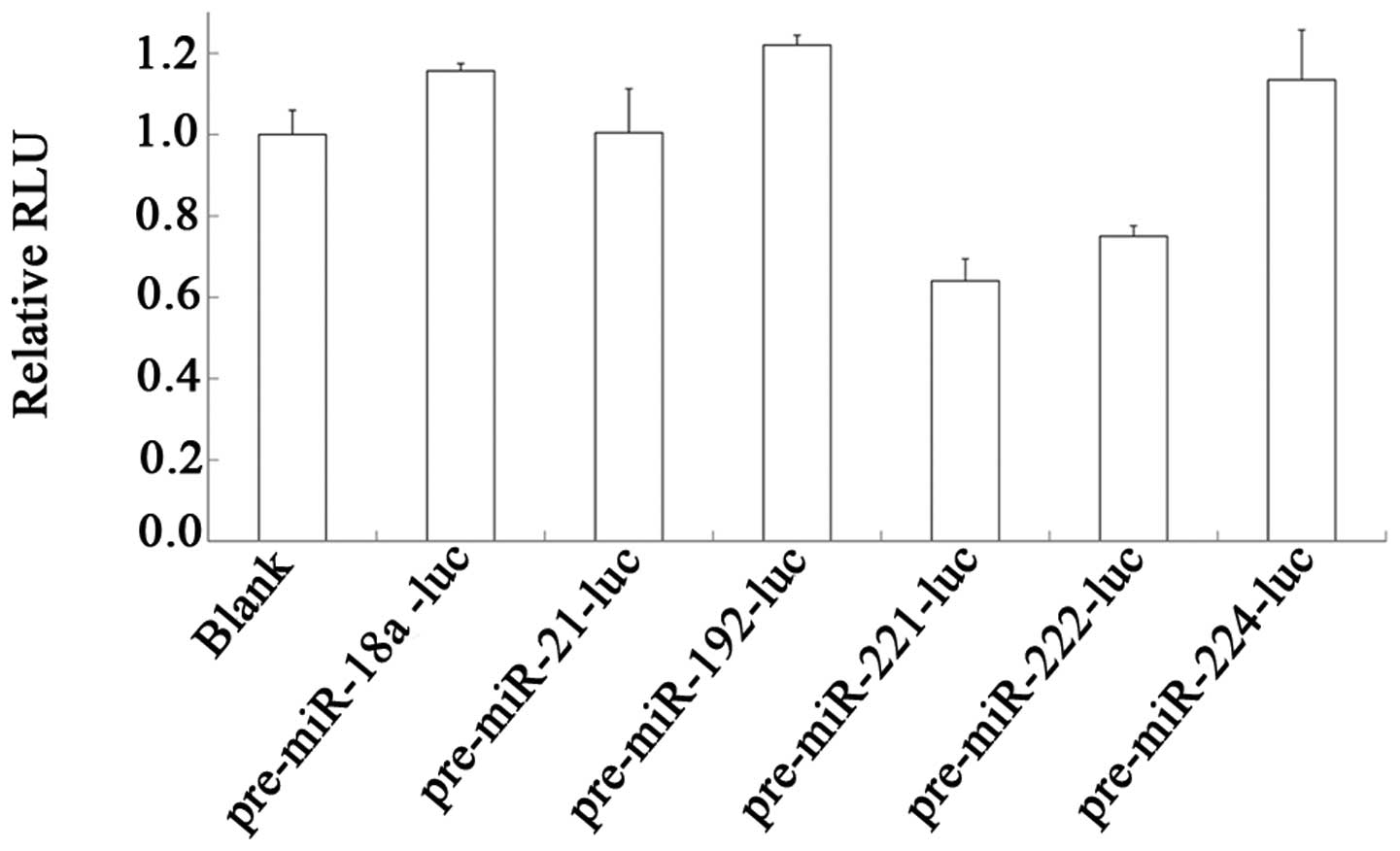
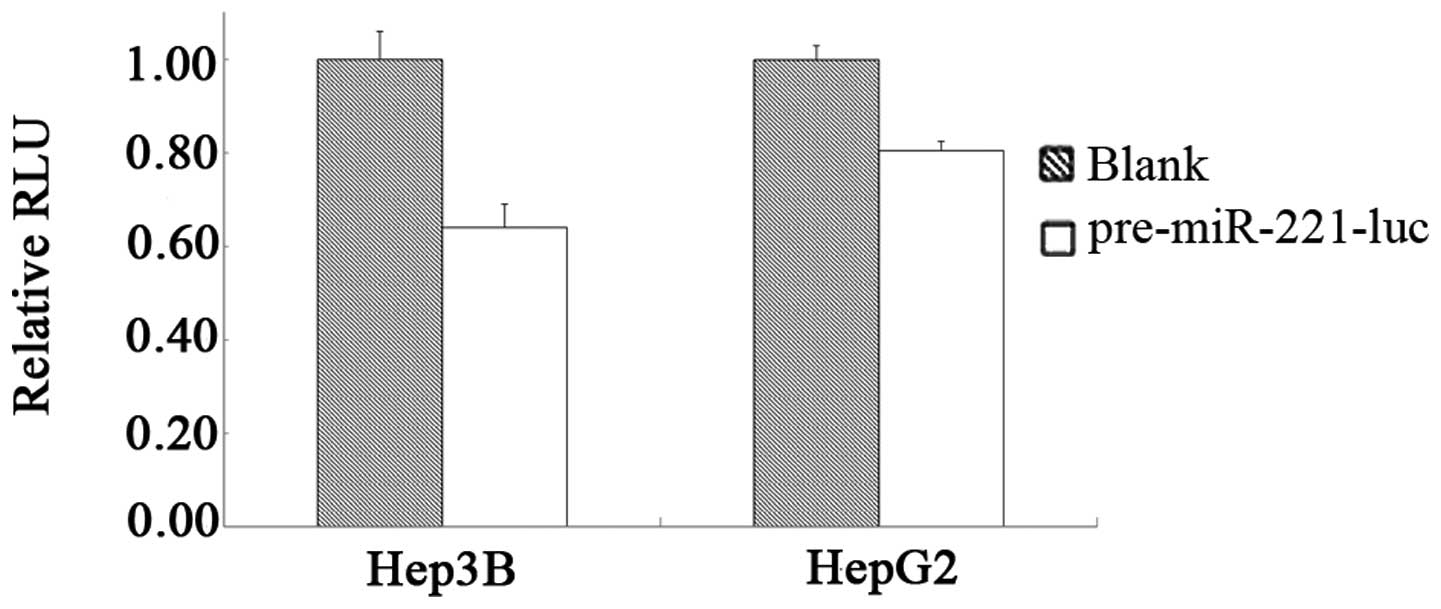
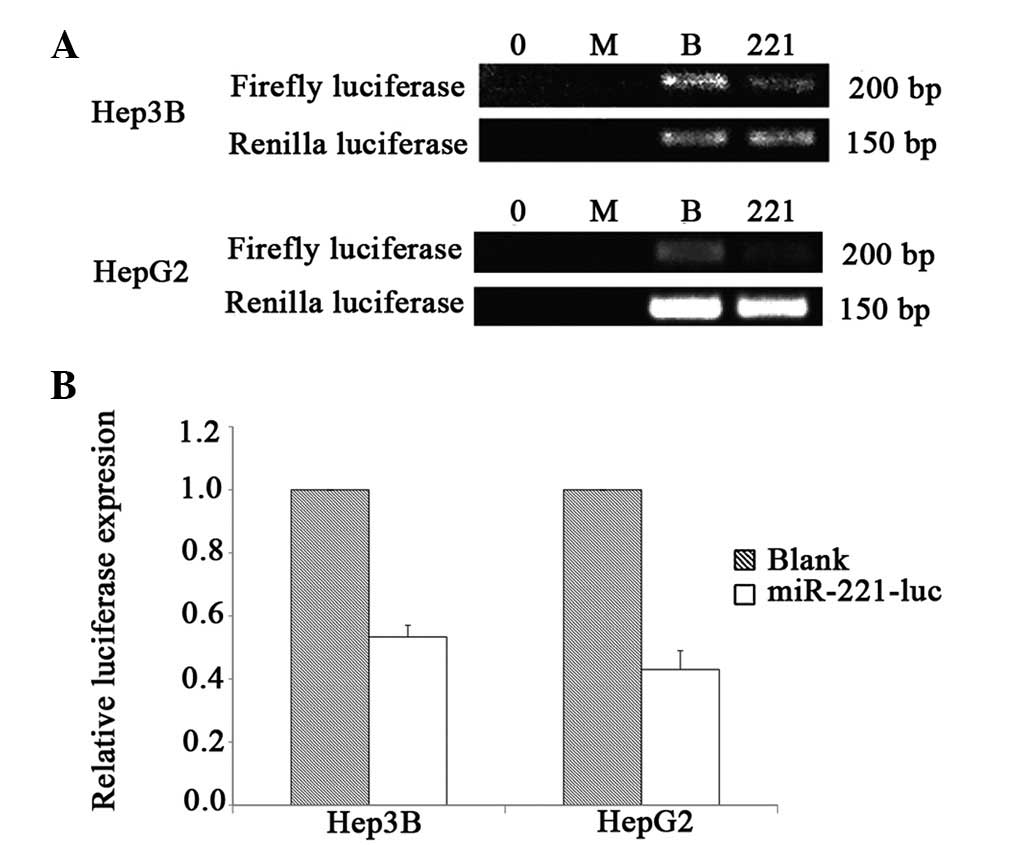
|
1.
|
Mathers C, Fat DM and Boerma J: The global
burden of disease: 2004 update. World Health Organization.
2008.
|
|
2.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Campbell TN and Choy FY: RNA interference:
past, present and future. Curr Issues Mol Biol. 7:1–6.
2005.PubMed/NCBI
|
|
4.
|
Sledz CA and Williams BRG: RNA
interference in biology and disease. Blood. 106:787–794. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Que XY, Li Y, Han Y and Li XZ: Effects of
siRNA-mediated Cdc2 silencing on MG63 cell proliferation and
apoptosis. Mol Med Rep. 7:466–470. 2013.PubMed/NCBI
|
|
6.
|
Yin Y, Chen X, Zhang CD, et al: Asymmetric
siRNA targeting the bcl-2 gene inhibits the proliferation of cancer
cells in vitro and in vivo. Int J Oncol. 42:253–260.
2013.PubMed/NCBI
|
|
7.
|
Boudreau RL, Martins I and Davidson BL:
Artificial microRNAs as siRNA shuttles: improved safety as compared
to shRNAs in vitro and in vivo. Mol Ther. 17:169–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liu X, Fang H, Chen H, et al: An
artificial miRNA against HPSE suppresses melanoma invasion
properties, correlating with a down-regulation of chemokines and
MAPK phosphorylation. PLoS One. 7:e386592012. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Slezak-Prochazka I, Durmus S, Kroesen BJ
and van den Berg A: MicroRNAs, macrocontrol: regulation of miRNA
processing. RNA. 16:1087–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Starega-Roslan J, Koscianska E, Kozlowski
P and Krzyzosiak WJ: The role of the precursor structure in the
biogenesis of microRNA. Cell Mol Life Sci. 68:2859–2871. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Zhang KJ, Qian J, Wang SB and Yang Y:
Targeting Gene-Viro-Therapy with AFP driving Apoptin gene shows
potent antitumor effect in hepatocarcinoma. J Biomed Sci.
19:202012. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Zhang KJ, Zhang J, Wu YM, et al: Complete
eradication of hepatomas using an oncolytic adenovirus containing
AFP promoter controlling E1A and an E1B deletion to drive IL-24
expression. Cancer Gene Ther. 19:619–629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Han J, Lee Y, Yeom KH, et al: Molecular
basis for the recognition of primary microRNAs by the Drosha-DGCR8
complex. Cell. 125:887–901. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Wang Y, Lee AT, Ma JZ, et al: Profiling
microRNA expression in hepatocellular carcinoma reveals
microRNA-224 up-regulation and apoptosis inhibitor-5 as a
microRNA-224-specific target. J Biol Chem. 283:13205–13215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Bala S, Marcos M and Szabo G: Emerging
role of microRNAs in liver diseases. World J Gastroenterol.
15:5633–5640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Boni V, Bandres E, Zarate R, Colucci G,
Maiello E and Garcia-Foncillas J: MicroRNAs as a new potential
therapeutic opportunity in gastrointestinal cancer. Oncology.
77(Suppl 1): 75–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Li M, Li J, Ding X, He M and Cheng SY:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar
|
|
18.
|
Ishida H, Tatsumi T, Hosui A, et al:
Alterations in microRNA expression profile in HCV-infected hepatoma
cells: involvement of miR-491 in regulation of HCV replication via
the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 412:92–97.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Albulescu R, Neagu M, Albulescu L and
Tanase C: Tissular and soluble miRNAs for diagnostic and therapy
improvement in digestive tract cancers. Expert Rev Mol Diagn.
11:101–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Silva JM, Li MZ, Chang K, et al:
Second-generation shRNA libraries covering the mouse and human
genomes. Nat Genet. 37:1281–1288. 2005.PubMed/NCBI
|
|
21.
|
Xu C, Lee SA and Chen X: RNA interference
as therapeutics for hepatocellular carcinoma. Recent Pat Anticancer
Drug Discov. 6:106–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Rutella S, Iudicone P, Bonanno G, et al:
Adoptive immunotherapy with cytokine-induced killer cells generated
with a new good manufacturing practice-grade protocol. Cytotherapy.
14:841–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Shojaei F: Anti-angiogenesis therapy in
cancer: current challenges and future perspectives. Cancer Lett.
320:130–137. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zeyaullah M, Patro M, Ahmad I, et al:
Oncolytic viruses in the treatment of cancer: a review of current
strategies. Pathol Oncol Res. 18:771–781. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Yao N, Yao D, Wang L, et al: Inhibition of
autocrine IGF-II on effect of human HepG2 cell proliferation and
angiogenesis factor expression. Tumour Biol. 33:1767–1776. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ibrahim AF, Weirauch U, Thomas M,
Grunweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Shi ZM, Wang J, Yan Z, et al: MiR-128
inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS
One. 7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Stegmeier F, Hu G, Rickles RJ, Hannon GJ
and Elledge SJ: A lentiviral microRNA-based system for single-copy
polymerase II-regulated RNA interference in mammalian cells. Proc
Natl Acad Sci USA. 102:13212–13217. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Chang K, Elledge SJ and Hannon GJ: Lessons
from Nature: microRNA-based shRNA libraries. Nat Methods.
3:707–714. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Hinton TM, Wise TG, Cottee PA and Doran
TJ: Native microRNA loop sequences can improve short hairpin RNA
processing for virus gene silencing in animal cells. J RNAi Gene
Silencing. 4:295–301. 2008.PubMed/NCBI
|
|
32.
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Augello C, Vaira V, Caruso L, et al:
MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster
as a novel prognostic biomarker in hepatocellular carcinoma. Liver
Int. 32:772–782. 2012. View Article : Google Scholar : PubMed/NCBI
|