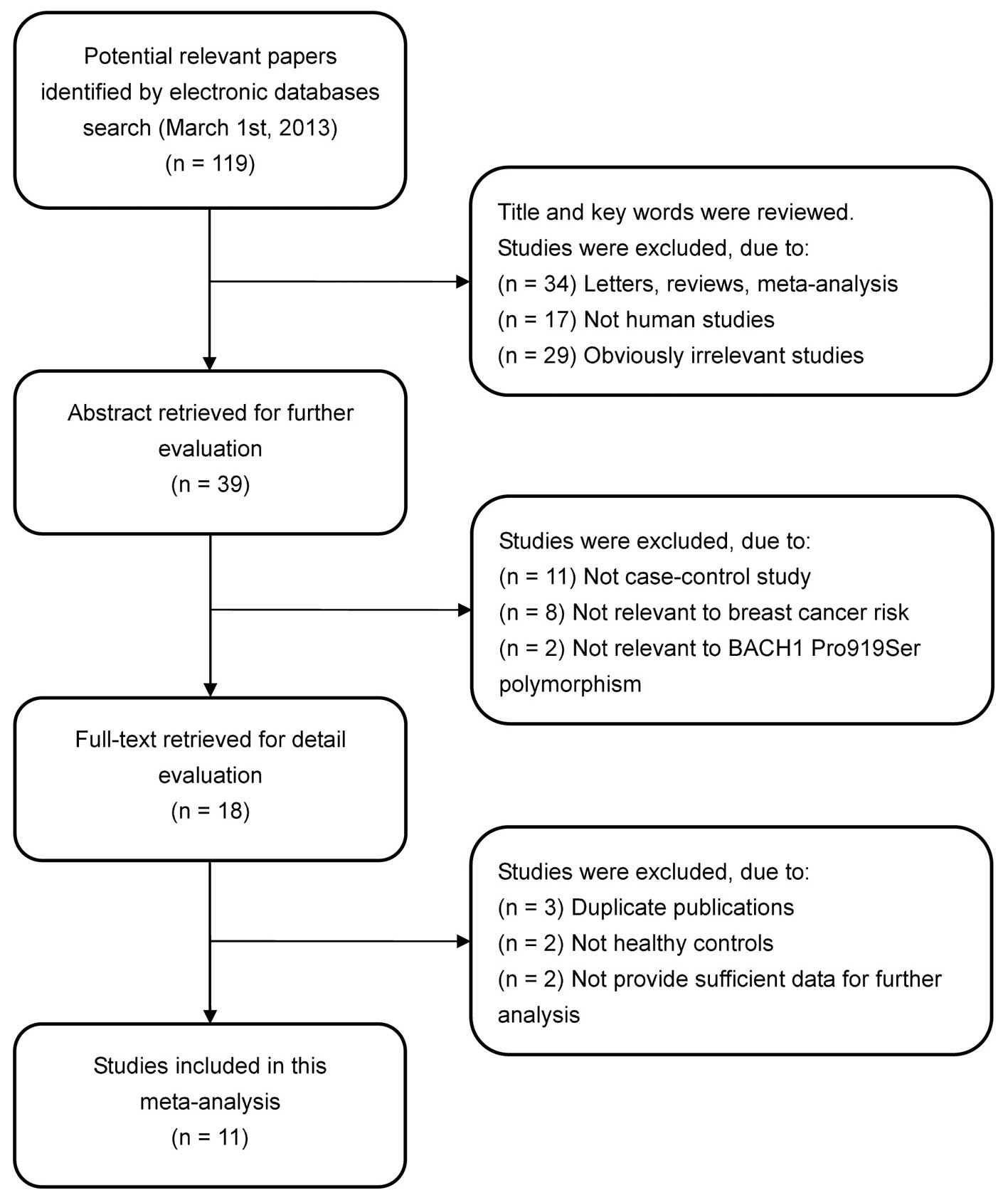
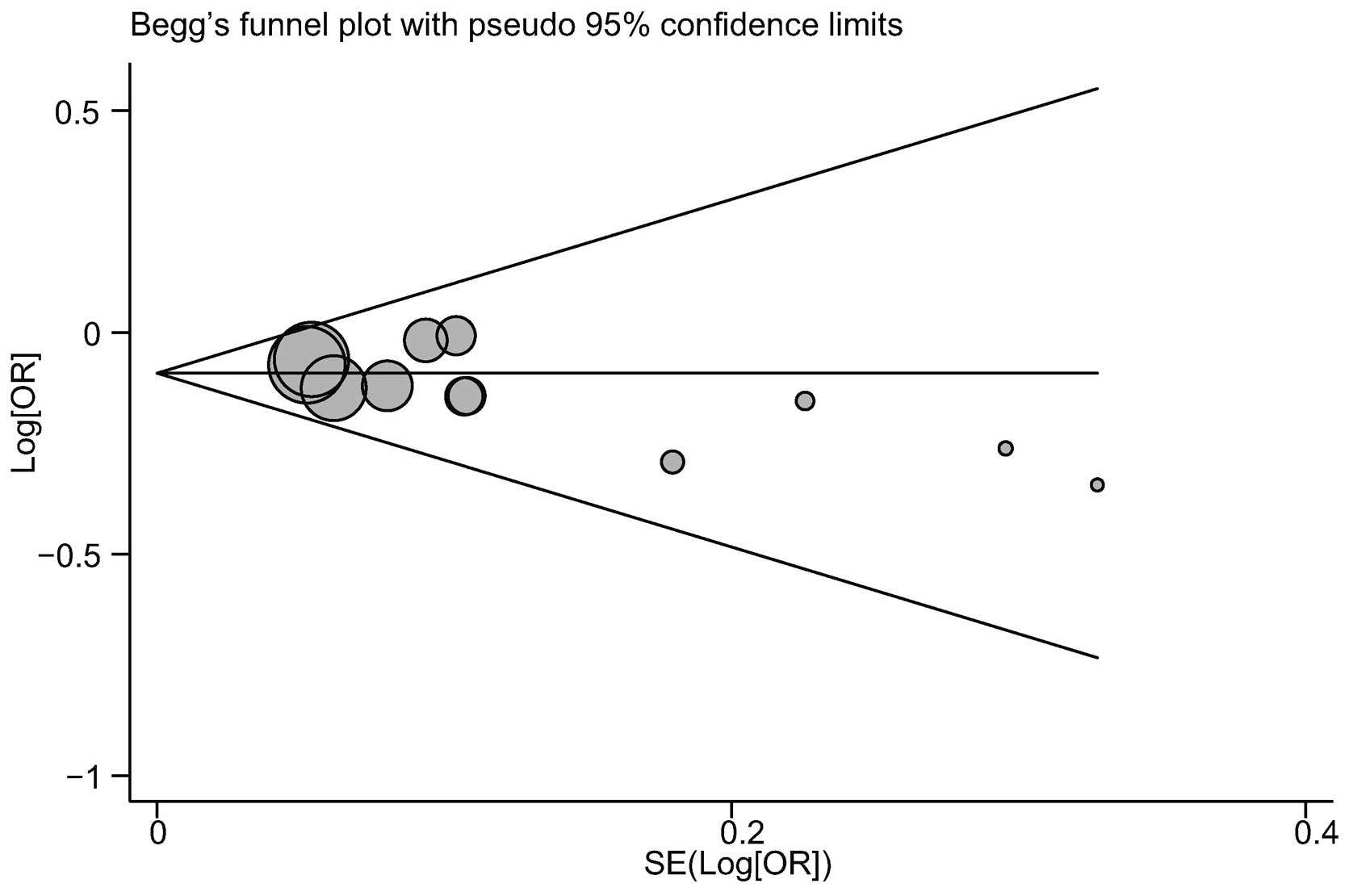
|
1.
|
Benson JR and Jatoi I: The global breast
cancer burden. Future Oncol. 8:697–702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4.
|
Wernberg JA, Yap J, Murekeyisoni C,
Mashtare T, Wilding GE and Kulkarni SA: Multiple primary tumors in
men with breast cancer diagnoses: a SEER database review. J Surg
Oncol. 99:16–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Prevalence and penetrance of BRCA1 and
BRCA2 mutations in a population-based series of breast cancer
cases. Anglian Breast Cancer Study Group. Br J Cancer.
83:1301–1308. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Pharoah PD, Tyrer J, Dunning AM, Easton DF
and Ponder BA; SEARCH Investigators: Association between common
variation in 120 candidate genes and breast cancer risk. PLoS
Genet. 3:e422007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kuusisto KM, Bebel A, Vihinen M,
Schleutker J and Sallinen SL: Screening for BRCA1, BRCA2, CHEK2,
PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish
BRCA1/2-founder mutation-negative breast and/or ovarian cancer
individuals. Breast Cancer Res. 13:R202011.PubMed/NCBI
|
|
8.
|
Cantor SB, Bell DW, Ganesan S, et al:
BACH1, a novel helicase-like protein, interacts directly with BRCA1
and contributes to its DNA repair function. Cell. 105:149–160.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Song H, Ramus SJ, Kjaer SK, et al: Tagging
single nucleotide polymorphisms in the BRIP1 gene and
susceptibility to breast and ovarian cancer. PLoS One. 2:e2682007.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Cantor SB and Guillemette S: Hereditary
breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future
Oncol. 7:253–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Vahteristo P, Yliannala K, Tamminen A,
Eerola H, Blomqvist C and Nevanlinna H: BACH1 Ser919Pro variant and
breast cancer risk. BMC Cancer. 6:192006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yu X, Chini CC, He M, Mer G and Chen J:
The BRCT domain is a phospho-protein binding domain. Science.
302:639–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Huo X, Lu C, Huang X, et al: Polymorphisms
in BRCA1, BRCA1-interacting genes and susceptibility of breast
cancer in Chinese women. J Cancer Res Clin Oncol. 135:1569–1575.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Huang J, Tang LL, Hu Z, et al: BRCA1 and
BRCA2 gene mutations of familial breast cancer and early-onset
breast cancer from Hunan Province in China. China Oncology.
18:566–572. 2008.
|
|
15.
|
Cao AY, Huang J, Hu Z, et al: Mutation
analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with
early onset breast cancer or affected relatives. Breast Cancer Res
Treat. 115:51–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Seal S, Thompson D, Renwick A, et al
Breast Cancer Susceptibility Collaboration (UK): Truncating
mutations in the Fanconi anemia J gene BRIP1 are low-penetrance
breast cancer susceptibility alleles. Nat Genet. 38:1239–1241.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Silvestri V, Rizzolo P, Falchetti M, et
al: Mutation analysis of BRIP1 in male breast cancer cases: a
population-based study in Central Italy. Breast Cancer Res Treat.
126:539–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Loizidou MA, Cariolou MA, Neuhausen SL, et
al: Genetic variation in genes interacting with BRCA1/2 and risk of
breast cancer in the Cypriot population. Breast Cancer Res Treat.
121:147–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Guénard F, Labrie Y, Ouellette G, Joly
Beauparlant C, Simard J and Durocher F; INHERIT BRCAs: Mutational
analysis of the breast cancer susceptibility gene BRIP1
/BACH1/FANCJ in high-risk non-BRCA1/BRCA2 breast cancer families. J
Hum Genet. 53:579–591. 2008.PubMed/NCBI
|
|
20.
|
Frank B, Hemminki K, Meindl A, et al:
BRIP1 (BACH1) variants and familial breast cancer risk: a
case-control study. BMC Cancer. 7:832007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
García-Closas M, Egan KM, Newcomb PA, et
al: Polymorphisms in DNA double-strand break repair genes and risk
of breast cancer: two population-based studies in USA and Poland,
and meta-analyses. Hum Genet. 119:376–388. 2006.PubMed/NCBI
|
|
22.
|
Rutter JL, Smith AM, Dávila MR, et al:
Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and
BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from
breast-ovarian cancer families and among early-onset breast cancer
cases and reference individuals. Hum Mutat. 22:121–128. 2003.
View Article : Google Scholar
|
|
23.
|
Pabalan N, Jarjanazi H and Ozcelik H:
Association between BRIP1 (BACH1) polymorphisms and breast cancer
risk: a meta-analysis. Breast Cancer Res Treat. 137:553–558. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Gallo V, Egger M, McCormack V, et al:
Strengthening the reporting of observational studies in
epidemiology - molecular epidemiology (STROBE-ME): an extension of
the STROBE Statement. PLoS Med. 8:e10011172011. View Article : Google Scholar
|
|
25.
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zintzaras E and Ioannidis JP:
Heterogeneity testing in meta-analysis of genome searches. Genet
Epidemiol. 28:123–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ioannidis JP, Patsopoulos NA and Rothstein
HR: Reasons or excuses for avoiding meta-analysis in forest plots.
BMJ. 336:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Peters JL, Sutton AJ, Jones DR, Abrams KR
and Rushton L: Comparison of two methods to detect publication bias
in meta-analysis. JAMA. 295:676–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Wong MW, Nordfors C, Mossman D, et al:
BRIP1, PALB2, and RAD51C mutation analysis reveals their relative
importance as genetic susceptibility factors for breast cancer.
Breast Cancer Res Treat. 127:853–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Cantor S, Drapkin R, Zhang F, et al: The
BRCA1-associated protein BACH1 is a DNA helicase targeted by
clinically relevant inactivating mutations. Proc Natl Acad Sci USA.
101:2357–2362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Rosenthal R and DiMatteo MR:
Meta-analysis: recent developments in quantitative methods for
literature reviews. Annu Rev Psychol. 52:59–82. 2001. View Article : Google Scholar
|
|
32.
|
Jüni P and Egger M: PRISMAtic reporting of
systematic reviews and meta-analyses. Lancet. 374:1221–1223.
2009.
|
|
33.
|
Ioannidis JP and Lau J: Pooling research
results: benefits and limitations of meta-analysis. Jt Comm J Qual
Improv. 25:462–469. 1999.PubMed/NCBI
|
|
34.
|
Dennis J, Hawken S, Krewski D, et al: Bias
in the case-only design applied to studies of gene-environment and
gene-gene interaction: a systematic review and meta-analysis. Int J
Epidemiol. 40:1329–1341. 2011. View Article : Google Scholar : PubMed/NCBI
|