Introduction
3-Methylindole (3MI) is a substance with an
unpleasant odor (1–4) that is produced by the tryptophan
removal process in the caecum and colon of intact male pigs
(5,6). It has been reported that consumers
are able to detect the negative odor when the level of 3MI is
>0.21 mg/kg (7).
In numerous countries, the castration of male
piglets is common practice to remove the odor. However, intact male
pigs have demonstrated a superior performance compared with
barrows, due to better carcass traits, lean meat percentage and
feed efficiency. Furthermore, the surgical castration of male
piglets not only diminishes the benefits of intact male pigs, but
also increases concerns about animal welfare (1). Surgical castration is not performed
in Australia, the United Kingdom and Ireland. In Norway, a total
ban on castration was initiated in January 2009 (8). However, if intact male pigs are to be
used for pork production, the 3MI level must be substantially
reduced. At present, a number of microbial methods have been used
to investigate the problem in vitro. Compared with physical
and chemical methods, biological techniques are preferable due to
the economical advantages and low possibility of byproduct
generation. Various bacterial strains have been applied to reduce
the levels of harmful substances for a number of years.
Ochrobactrum intermedium DN2 has been used to degrade
nicotine in tobacco waste extracts and the average degradation rate
of nicotine in a 30 l fed-batch culture was 140.5 mg/l/h (9). In addition, Shinella
zoogloeoides BC026 has been identified to reduce pyridine
levels, resulting in a degradation rate of 1,806 mg/l pyridine in
45.5 h (10). Moreover,
Bacillus odysseyi SUK3, Morganella morganii SUK5 and
Proteus species SUK7 have been shown to decolorize Reactive
Blue 59 (50 mg/l) completely within 60, 30 and 24 h, respectively
(11). With regard to 3MI
degradation, Kohda et al (12) identified that 0.05% (w/v) 3MI may
be degraded by a type of Clostridium from the feces of pigs
in under 4 weeks with a removal rate of up to 32.18%. Yin et
al (13) demonstrated that 2.5
mmol/l 3MI may be reduced by Pseudomonas aeruginosa
(extracted from the sediment of lapacho wood) in 3 days and the
time was extended as the 3MI concentration increased from 2.5 to
3.5 mmol/l. Additionally, Gu and Berry (14) indicated that 1–1.5 μmol/l
3MI was reduced in 36 days by a bacterial colony that produced
methane. Furthermore, Gu et al (15) reported that 3MI may be completely
degraded by sulfate-reducing bacteria.
The objectives of the present study were to
investigate the growth of lactic acid bacteria [Lactobacillus
brevis 1.12 (L. brevis 1.12), L. plantarum 102,
L. casei 6103 and L. plantarum ATCC8014] in culture
medium with varied concentrations of 3MI, and to explore the
correlation between the levels of 3MI and the 3MI removal ability
of the lactic acid bacteria during the fermentation process in
vitro.
Materials and methods
Strains and medium
L. brevis 1.12, L. casei 6103 and
L. plantarum ATCC8014 were purchased from China Center of
Industrial Culture Collection (Beijing, China). L. plantarum
102 was obtained from American Type Culture Collection (Manassas,
VA, USA). MRS broth (Oxoid Ltd., Basingstoke, UK) was used as the
culture medium for the lactic acid bacteria.
Chemicals and reagents
3MI, acetonitrile and methanol of high-performance
liquid chromatography (HPLC) grade were purchased from
Sigma-Aldrich (St. Louis, MO, USA). Phosphate-buffered saline (PBS;
Sigma-Aldrich) was of analytical grade. All remaining chemicals
were of biological and analytical reagent grades, and were obtained
from Kelong Chemical Reagent Factory (Chengdu, China).
Effects of 3MI on the growth of lactic
acid bacteria
The 3MI standard solutions contained 0.0, 0.2, 0.4,
0.6, 0.8 and 1.0 g 3MI standard substance dissolved in 10 ml
absolute ethyl alcohol (w/v; 0.00, 0.02, 0.04, 0.06, 0.08 and 0.10
g/ml, respectively). The bacterial colonies of L. brevis
1.12, L. plantarum 102, L. casei 6103 and L.
plantarum ATCC8014 were suspended in 10 ml 0.75% (w/v)
physiological saline. Subsequently, 2.5% (v/v) suspension liquid
and 1 ml 3MI solution were mixed with 100 ml MRS medium for 72 h at
37°C. The optical density (OD) value at 600 nm was detected every 2
h by a spectrophotometer (722-spectrophotometer; Tairui Instrument
Co., Ltd., Chongqing, China) (16,17).
Removal of 3MI from the MRS broth by
fermentation of lactic acid bacteria strains
3MI standard substance (0.0, 0.2, 0.4, 0.6, 0.8 and
1.0 g) was dissolved in 10 ml absolute ethyl alcohol to provide 3MI
standard solutions (0.00, 0.02, 0.04, 0.06, 0.08 and 0.10 g/ml,
respectively). Bacterial colonies of L. brevis 1.12, L.
plantarum 102, L. casei 6103 and L. plantarum
ATCC8014 were suspended in 10 ml 0.75% (w/v) physiological saline.
3MI solution (1 ml) and 2.5% (v/v) suspension liquid were mixed in
100 ml MRS medium for 120 h at 37°C. The sample treatment method
for HPLC was as follows: The fermentation broth was centrifuged at
9000 × g for 10 min; 1 ml of supernatant was mixed with 9 ml
acetonitrile:ultrapure water (75:25, v/v); and the intermixture was
filtered through an organic phase filter of 0.45 μm
(Frisenette ApS Co., Ebeltoft, Denmark) and loaded into a 1.5 ml
screw-thread bottle (16,17). In the present study, the removal
rate of 3MI (%) was the response value, which was calculated using
the following equation: Removal rate (%) = (A-B)/Ax100; where A is
the initial level of 3MI (ng/ml) and B represents the residual
level of 3MI (ng/ml).
Removal of 3MI by the supernatant fluid
of fermentation, cell pellets and extracts of lactic acid
bacteria
An activated culture of lactic acid bacteria (10 ml)
was centrifuged (9,000 × g, 5 min, 5°C). The supernatant fluid of
fermentation was collected, and the resultant cell pellets were
washed twice with 10 ml sterile PBS (0.01 M, pH 7.4) and suspended
in 10 ml sterile PBS. The cell pellet suspension (10 ml) was
disintegrated (400 W every 5 sec for 30 min) in an ice-water bath
by an ultrasonic cell disintegrator (Branson Sonifier 450; Branson
Ultrasonics Corp., Danbury, CT, USA). The disintegrated cell
suspension was centrifuged (9,000 × g, 5 min, 5°C) and the
supernatant (cell extract) was collected. 3MI was added to the
solutions of supernatant fluid of fermentation, suspensions of cell
pellets and cell extracts of lactic acid bacteria, to an initial
3MI concentration of 1.0 μg/ml. Sterile PBS containing 1.0
μg/ml 3MI was used as the control. The suspensions were
incubated at 37°C for 24 h, centrifuged (9,000 × g, 5 min, 5°C) and
the supernatant fluids were filtered through a 0.45-μm
filter and stored at 4°C prior to analysis (18,19).
Removal of 3MI from PBS by viable heat-,
acid- and alkali-treated cells and the supernatant fluid of lactic
acid bacteria
Activated culture of L. brevis 1.12 (10 ml)
was centrifuged (9,000 × g, 5 min, 5°C) and the cell pellets were
washed twice with 10 ml sterile PBS (0.01 M, pH 7.4). Cells of
L. brevis 1.12 were treated by the following methods:
heating (100°C for 30 min, incubated at 37°C for 24 h), acid
treatment (1 M HCl, incubated at 37°C for 24 h) and alkali
treatment (1 M NaOH, incubated at 37°C for 24 h). Following these
treatments, the suspensions were centrifuged (9,000 × g, 5 min,
5°C) and the supernatants were removed. The resultant cell pellets
were washed twice with 10 ml sterile PBS and then suspended in 10
ml sterile PBS. Sterile PBS containing 1.0 μg/ml 3MI was
used as the control. The removal of 3MI was tested as previously
described. The supernatant fluid of fermentation was cryodesiccated
and treated by the same method used for cells of L. brevis
1.12 (20,21).
Analysis of 3MI by HPLC
For HPLC, an LC-20A system (Shimadzu Co., Kyoto,
Japan) was used consisting of a SIL-10ADvp injector with a 100
μl loop and two LC-10 ADvp HPLC pumps. The detector used was
a RF-20A fluorometer and data were collected with an LC solution
integrator. The column was Hypersil (ODS-2, 5-μm particle
size; length, 200 mm; tubing I.D, 4.6 mm; Elite Analytical
Instrument Co., Ltd., Dalian, China) and operated at ambient
temperature. The mobile phase consisted of acetonitrile:ultrapure
water (60:40, v/v) and the flow rate was 1.0 ml/min. The detection
was carried out by measuring the fluorescence with the following
wavelengths: excitation at 281 nm and emission at 353 nm. The
volume of the injected sample was 10 μl (22).
Statistical analysis
All data are expressed as the mean ± standard
deviation of triplicate assays. Simple linear regressions for 3MI
standard solutions with different gradient concentrations and the
growth of lactic acid bacteria in the MRS medium with 3MI were
calculated using Microsoft Excel 2007. Statistical analyses for the
removal rate of 3MI (%) were carried out using PASW statistics
(formerly SPSS), version 18.0 (IBM SPSS, Inc., Chicago, IL,
USA).
Results
Effects of 3MI on the growth of lactic
acid bacteria
The growth responses of lactic acid bacteria (L.
brevis 1.12, L. plantarum 102, L. casei 6103 and
L. plantarum ATCC8014) to different concentrations of 3MI
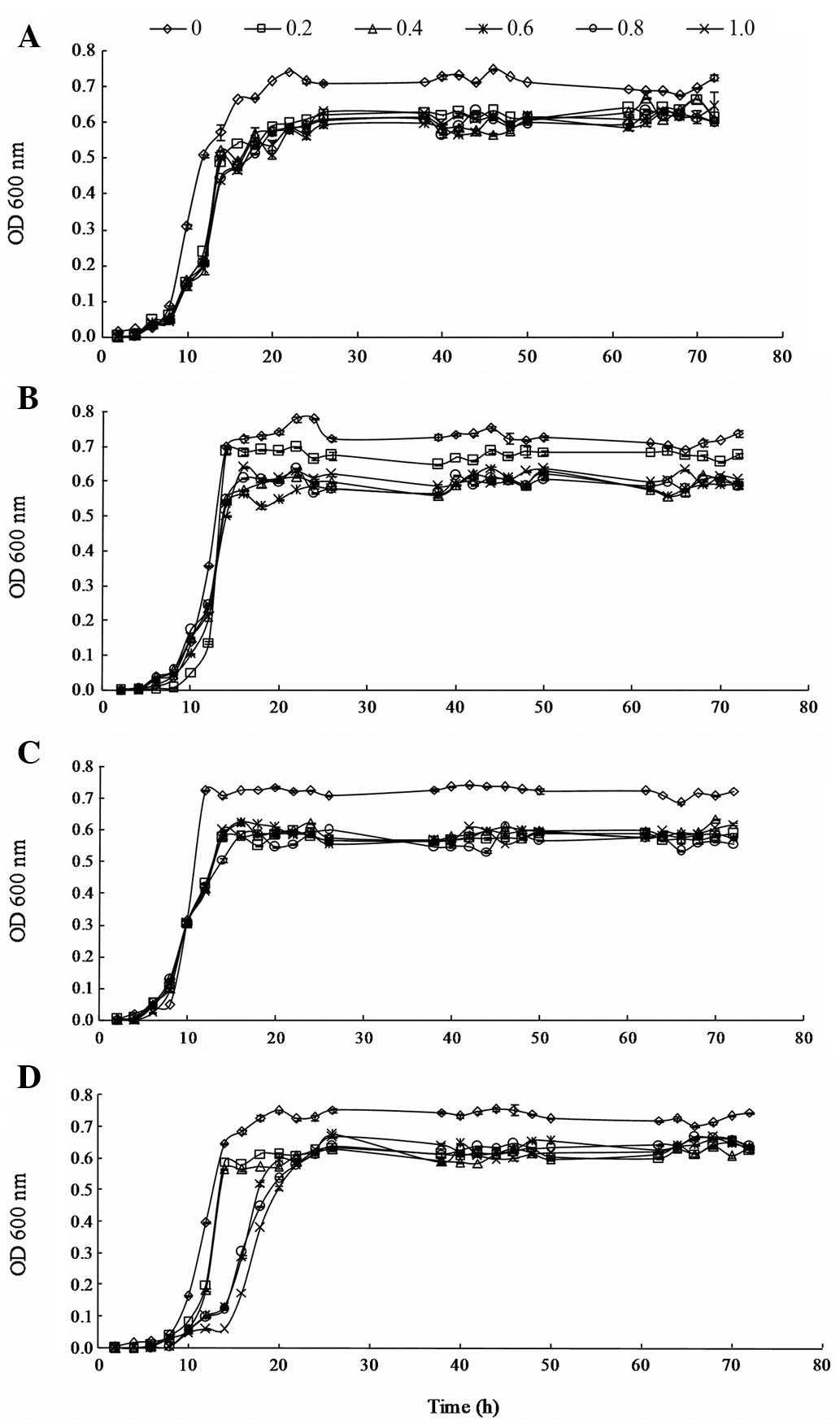
(from 0.2 to 1.0 μg/ml) are shown in Fig. 1.
The effects of 3MI on the growth of L. brevis
1.12 were not significant during the lag phase. However, the growth
of L. brevis 1.12 was inhibited by 3MI during the
logarithmic phase. During the stationary phase, the OD values of
L. brevis 1.12 in the MRS medium containing 3MI were lower
than such values of L. brevis 1.12 in MRS medium without
3MI. However, the growth of L. brevis 1.12 was steady as the
level of 3MI increased from 0.2 to 1.0 μg/ml in the
stationary phase.
The effects of 3MI on the growth of L.
plantarum 102 were not significant at the lag and logarithmic
phases. In addition, the results showed that the growth of L.
plantarum 102 was not inhibited by 3MI at the two phases.
During the stationary phase, the growth of L. plantarum 102
was restrained in the presence of increasing levels of 3MI (from
0.2 to 1.0 μg/ml) and significant differences between the
effects of the various levels of 3MI on the growth of L.
plantarum 102 were observed.
The effects of 3MI on the growth of L. casei
6103 were not significant at the lag and logarithmic phases;
however, growth was slower than that of the control group at 10–12
h in the logarithmic phase. At the stationary phase, the growth of
L. casei 6103 was restrained in the MRS medium with 0.2
μg/ml 3MI, but the OD value was steady as the level of 3MI
increased from 0.2 to 1.0 μg/ml. The results suggest that
the growth of L. casei 6103 was inhibited by 3MI at the
stationary phase; however, the difference in growth among the
varied levels of 3MI was not marked.
The effects of 3MI on the growth of L.
plantarum ATCC8014 were significant at the logarithmic phase,
with slower growth compared with that of the control. However,
during the stationary phase, the growth was steady with increasing
concentrations of 3MI from 0.2 to 1.0 μg/ml.
3MI removal during the fermentation of
lactic acid bacteria
The relationship between the concentration of 3MI
and the removal ability of the four strains during the fermentation
process was also studied. The results indicate that the levels of
3MI decreased during the fermentation process in all four strains
and the removal rate increased as the incubation time increased
from 24 to 120 h. Differences among the four strains were
significant, as shown in Table I.
L. brevis 1.12 indicated the strongest ability to remove 3MI
compared with the other strains. The removal rate increased as the
incubation time was extended from 24 to 120 h; however, the effects
of different 3MI levels on the ability of L. brevis 1.12 to
remove MI were not significant, as the removal rate was steady with
increasing levels of 3MI from 0.2 to 1.0 μg/ml; the highest
removal rate was 65.35±0.3% in the fermentation fluid of L.
brevis 1.12 with 1.0 μg/ml 3MI in 120 h. For L.
plantarum 102, L. casei 6103 and L. plantarum
ATCC8014, the effects of the different levels of 3MI on the removal
ability of the three strains were significant. The ability of the
three strains to remove 3MI decreased as the 3MI levels increased
from 0.2 to 1.0 μg/ml, and L. plantarum 102 and L.
plantarum ATCC8014 were more sensitive to 3MI when compared
with L. casei 6103. The removal ability of L.
plantarum 102 and L. plantarum ATCC8014 was sensitive to
0.4 and 0.8 μg/ml 3MI, respectively. The removal rates were
28.54±0.2 and 33.23±0.9% in the fermentation fluid of L.
plantarum 102 and L. plantarum ATCC8014 with 1.0
μg/ml 3MI, respectively, in 120 h.
 | Table I.Effects of the level of 3MI on the
3MI removal ability of lactic acid bacteria. |
Table I.
Effects of the level of 3MI on the
3MI removal ability of lactic acid bacteria.
| Concentration of
3MI (μg/ml) | Incubation time
(h) | Removal rate (%)
|
|---|
| L. brevis
1.12 | L. plantarum
102 | L. casei
6103 | L. plantarum
ATCC8014 |
|---|
| 0.2 | 24 |
15.71±0.20a |
11.47±0.40b |
9.20±0.10c |
11.55±0.20b |
| 48 |
30.62±0.10b |
39.77±0.10a |
21.39±0.40c |
30.50±0.10b |
| 72 |
57.36±0.60a |
53.33±0.30c |
54.23±1.50b |
50.27±0.30d |
| 96 |
62.64±0.10a |
57.28±0.10c |
60.30±0.20b |
52.09±0.10d |
| 120 |
67.63±0.20a |
60.79±0.10c |
61.53±1.20b |
56.75±0.10d |
| 0.4 | 24 |
15.36±0.10a |
9.46±0.30b |
6.28±0.60c |
9.50±0.20b |
| 48 |
29.81±0.10a |
25.38±0.10b |
20.72±0.50c |
17.90±0.20d |
| 72 |
57.20±0.10a |
46.21±0.50c |
53.55±0.40b |
26.64±0.90d |
| 96 |
62.12±0.10a |
53.51±0.50c |
57.14±0.20b |
27.32±0.20d |
| 120 |
67.87±0.20a |
55.02±0.70c |
59.16±0.10b |
35.41±0.30d |
| 0.6 | 24 |
15.13±0.60a |
2.65±0.40d |
6.21±0.50c |
7.70±0.80b |
| 48 |
30.93±0.20a |
19.38±0.30c |
20.16±0.50b |
17.93±0.10d |
| 72 |
57.52±0.20a |
40.23±0.60c |
46.89±0.60b |
23.96±0.10d |
| 96 |
61.76±0.40a |
46.72±0.10c |
50.32±0.60b |
26.73±0.70d |
| 120 |
67.61±0.30a |
47.51±0.20c |
59.14±0.20b |
35.20±0.20d |
| 0.8 | 24 |
16.09±0.10a |
1.05±0.10d |
4.21±0.50c |
8.12±0.30b |
| 48 |
31.23±0.20a |
12.44±0.50d |
17.21±0.80b |
15.53±0.40c |
| 72 |
57.66±0.20a |
33.35±0.30c |
46.40±0.40b |
22.35±0.10d |
| 96 |
61.63±0.10a |
35.64±0.20c |
52.18±0.60b |
23.27±0.60d |
| 120 |
67.12±0.10a |
35.68±0.30c |
52.76±0.10b |
35.71±0.10c |
| 1.0 | 24 |
17.43±0.20a |
0.88±0.10d |
3.48±0.26c |
5.50±0.40b |
| 48 |
31.68±0.20a |
10.62±0.20d |
16.43±0.20b |
19.05±0.90c |
| 72 |
56.72±0.60a |
29.60±0.20c |
39.32±0.30b |
25.42±0.50d |
| 96 |
61.55±0.60a |
29.63±0.10c |
51.78±0.20b |
26.16±0.50d |
| 120 |
65.35±0.30a |
28.54±0.20d |
52.15±0.30b |
33.23±0.90c |
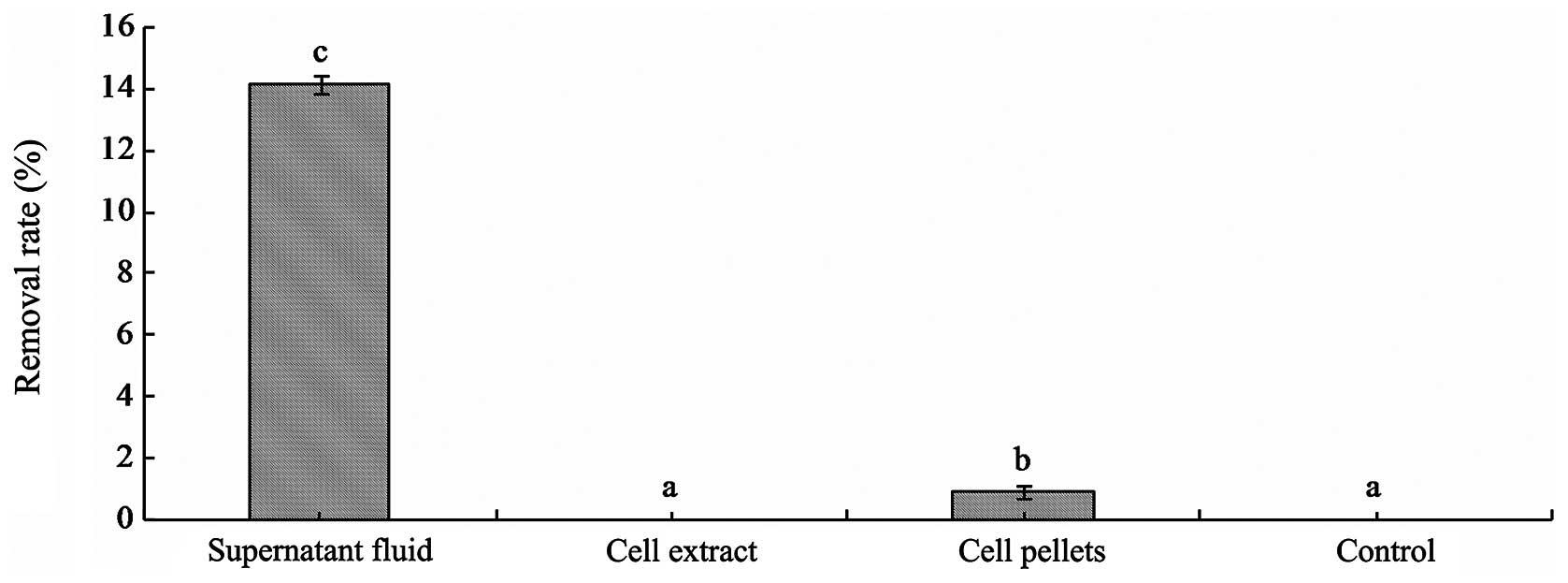
Mode of removal
The concentrations of 3MI following incubation with
the supernatant fluid of fermentation, the suspension of cell
pellets and cell extracts of L. brevis 1.12 at 37°C for 24 h
were detected by HPLC. The results showed that the 3MI removal
ability of the supernatant fluid of fermentation broth was the
strongest. 3MI was removed by the supernatant fluid of fermentation
with a removal rate of 14.4±0.3% at 37°C for 24 h. 3MI was removed
by cell pellets of L. brevis 1.12 (0.88±0.2%) (Fig. 2), but 3MI was not detected in the
PBS eluent. The results suggest that the removal mode of 3MI was
not through the physical binding of cells by L. brevis 1.12.
Furthermore, the results showed that the removal rates of 3MI in
the suspensions following incubation with heat-, acid- and
alkali-treated cells decreased significantly, and the removal
ability of L. brevis 1.12 was inhibited under these methods
(Table II). The results also
confirmed that the removal mode for 3MI was not via physical
binding.
 | Table II.Effects of supernatant and cell
pellets of L. brevis 1.12 on 3MI removal using different
treatment methods. |
Table II.
Effects of supernatant and cell
pellets of L. brevis 1.12 on 3MI removal using different
treatment methods.
| Substance | Removal rate (%)
|
|---|
| Heat-treated | Acid-treated | Alkali-treated | Non-treated | HCl control | NaOH control |
|---|
| Supernatant |
3.98±1.3a |
6.70±0.7c |
4.92±1.2b |
15.27±2.3d | − | − |
| Cell pellets | − | − | − | 0.95±1.5 | − | − |
Discussion
In the present study, the results suggest that the
four strains of lactic acid bacteria are more sensitive to 3MI than
previously investigated microorganisms from other studies regarding
the effects of 3MI on microorganisms. The growth of all four
strains was inhibited by low levels of 3MI (0.2 μg/ml).
However, Dreizen and Spies (23)
identified that the growth of L. acidophilus was completely
restricted by 400 μg/ml 3MI, but growth occurred when the
3MI concentration was decreased from 400 to 100 μg/ml. In
addition, Tittlser et al (24) demonstrated that the growth of 25
species of Gram-negative bacteria extracted from the intestinal
tract was inhibited when the 3MI concentration was 330
μg/ml. Furthermore, Kohda et al (12) identified that the growth of
Clostridium was steady in 100–300 μg/ml 3MI solution;
however, the growth of certain clostridia was prevented in 50
μg/ml 3MI solution. In the present study, the growth of the
four strains was prevented in the incubation environment with 0.2
μg/ml 3MI and the tolerance levels for 3MI concentration
were lower than those identified in the previously mentioned
studies.
Furthermore, the ability of the four strains to
degrade 3MI was compared with results from previous studies. The
results showed that the 3MI removal ability of the four strains was
stronger than that of the microorganisms investigated in the
studies by Gu et al (15)
and Li et al (25). Gu
et al (15) identified that
3MI may be degraded by marine anaerobic microorganisms for 30 days.
Additionally, Li et al (22) indicated that 2.0 mmol/l 3MI may be
degraded by Pseudomonas putida LPC24 in <30 days. In the
present study, the degradation ability of L. brevis 1.12 was
the strongest among the bacteria tested, with a degradation rate
for 1.0 μg/ml 3MI of 65.35±0.3% in 5 days. However, the
degradation time was longer than that in the study by Yin et
al (13). The study suggested
that 2.5 mmol/l 3MI may be reduced by Pseudomonas aeruginosa
(extracted from the sediment of lapacho wood) in 3 days and the
time extended with increased 3MI concentration from 2.5 to 3.5
mmol/l (13). The different
results may be due to significant differences among the
microorganisms tested.
The results of the present study demonstrated that
3MI may be degraded by the supernatant fluid of fermentation and
suspension of cell pellets; however, 3MI was not detected in the
eluent of cell pellets. This suggests the key substance responsible
for the degradation of 3MI exists in the supernatant fluid of the
fermentation broth and that the mode of 3MI removal was not through
the physical binding of cells by L. brevis 1.12. However, in
the present study, the removal mechanism of 3MI during the
fermentation process of L. brevis 1.12 was not studied. The
removal mechanism of 3MI in certain microorganisms and the liver of
entire pigs have been demonstrated in previous studies. Gu et
al (15) identified that 3MI
may be degraded by marine anaerobic microorganisms, and the
mechanism included two steps of oxidation accomplished by
hydroxylation and then dehydrogenation at the 2- and 3-positions
sequentially prior to the cleavage of the pyrrole ring between the
2- and 3-positions. The 3MI degradation mechanism in pig liver is
usually conducted in two steps, phase I and II (26). Phase I consists of an oxidation of
the compound, usually catalyzed by cytochrome P450 (CYP450)
enzymes, while phase II is conducted by a more diverse group of
enzymes and consists of conjugation with a hydrophilic group, such
as by glucuronidation, sulfoconjugation or glucosidation. The
outcome of phase I and/or II metabolism is often the elimination of
the compound by excretion (26).
In pigs, the phase I metabolism of 3MI is mainly mediated by
hepatic CYP1A2, CYP2A and CYP2E1 (27). Diaz and Squires (28) indicated that 3MI is metabolized by
the CYP450 system in the lungs and liver of ruminants, rodents and
humans. Squires and Lundström (26) demonstrated that a similar system
may be involved in 3MI metabolism in pigs and a particular CYP2E1
was indicated as the major enzyme responsible for metabolic
breakdown of 3MI in the liver. Moreover, Chen et al
(8) identified that a dietary
supplement of raw potato starch reduced the levels of 3MI.
Furthermore, the addition of fructooligosaccharide to pig fecal
slurries significantly reduced 3MI levels but not indole synthesis
from tryptophan. In future studies, the supernatant fluid of the
fermentation broth with 3MI will be detected using HPLC to analyze
whether new metabolites are produced and to study the mechanism of
3MI degradation by L. brevis 1.12.
In conclusion, the present study used various in
vitro experimental methods to investigate the growth of lactic
acid bacteria in the presence of 3MI, the 3MI removal ability of
four strains of bacteria during the fermentation process and the
removal mode of 3MI by L. brevis 1.12. The results
demonstrated that the growth of all four strains was inhibited by
3MI and the 3MI removal ability of L. brevis 1.12 was the
strongest. The 3MI removal rate of L. brevis 1.12 was
65.35±0.3% from the incubation medium of 1 ml 1.0 μg/ml 3MI
in 120 h. Furthermore, this study demonstrated that the mode of 3MI
removal was not through the physical binding of cells by L.
brevis 1.12.
Acknowledgements
This study was funded by the National
Basic Research Program of China (973 program; grant no.
2009CB118806), the National Rabbit Industry Technology System
Program (grant no. CARS-44D-1) and the National Natural Science
Foundation of China (grant no. 31071566).
References
|
1.
|
Barton Gade PA: Meat and fat quality in
boars, castrates and gilts. Livest Prod Sci. 16:187–196. 1987.
|
|
2.
|
Bonneau M, Dufour R, Chouvet C, Roulet C,
Meadus W and Squires EJ: The effects of immunization against
luteinizing hormone-releasing hormone on performance, sexual
development, and levels of boar taint-related compounds in intact
male pigs. J Anim Sci. 72:14–20. 1994.
|
|
3.
|
Babol J, Squires EJ and Lundström K:
Relationship between metabolism of androstenone and skatole in
intact male pigs. J Anim Sci. 77:84–92. 1999.PubMed/NCBI
|
|
4.
|
Babol J, Squires EJ and Lundström K:
Hepatic metabolism of skatole in pigs by cytochrome P4502E1. J Anim
Sci. 76:822–828. 1998.PubMed/NCBI
|
|
5.
|
Jensen MT, Cox RP and Jensen BB:
3-Methylindole (skatole) and indole production by mixed populations
of pig fecal bacteria. Appl Environ Microbiol. 61:3180–3184.
1995.PubMed/NCBI
|
|
6.
|
Yokoyama MT and Carlson JR: Microbial
metabolites of tryptophan in the intestinal tract with special
reference to skatole. Am J Clin Nutr. 32:173–178. 1979.PubMed/NCBI
|
|
7.
|
Lunde K, Skuterud E, Hersleth M and
Egelandsdal B: Norwegian consumers’ acceptability of boar tainted
meat with different levels of androstenone or skatole as related to
their androstenone sensitivity. Meat Sci. 86:706–711. 2010.
|
|
8.
|
Chen G, Zamaratskaia G, Andersson HK and
Lundström K: Effects of raw potato starch and live weight on fat
and plasma skatole, indole and androstenone levels measured by
different methods in entire male pigs. Food Chem. 101:439–448.
2007. View Article : Google Scholar
|
|
9.
|
Yuan YJ, Lu ZX, Huang LJ, Li Y, Lu FX, Bie
XM, et al: Biodegradation of nicotine from tobacco waste extract by
Ochrobactrum intermedium DN2. J Ind Microbiol Biotechnol.
34:567–570. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Bai YH, Sun Q, Zhao C, Wen D and Tang X:
Aerobic degradation of pyridine by a new bacterial strain,
Shinella zoogloeoides BC026. J Ind Microbiol Biotechnol.
36:1391–1400. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Patil PS, Shedbalkar UU, Kalyani DC and
Jadhav JP: Biodegradation of Reactive Blue 59 by isolated bacterial
consortium PMB11. J Ind Microbiol Biotechnol. 35:1181–1190. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Kohda C, Ando T and Nakai Y: Isolation and
characterization of anaerobic indole- and skatole-degrading
bacteria from composting animal wastes. J Gen Appl Microbiol.
43:249–255. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yin B, Huang L and Gu JD: Biodegradation
of 1-methylindole and 3-methylindole by mangrove sediment
enrichment cultures and a pure culture of an isolated
Pseudomonas aeruginosa Gs. Water Air Soil Poll. 176:185–199.
2006. View Article : Google Scholar
|
|
14.
|
Gu JD and Berry DF: Metabolism of
3-methylindole by a methanogenic consortium. Appl Environ
Microbiol. 58:2667–2669. 1992.PubMed/NCBI
|
|
15.
|
Gu JD, Fan Y and Shi H: Relationship
between structures of substituted indolic compounds and their
removal by marine anaerobic microorganisms. Mar Pollut Bull.
45:379–384. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
El-Nezami HS, Polychronaki N, Salminen S
and Mykkänen H: Binding rather than metabolism may explain the
interaction of two food-grade Lactobacillus strains with
zearalenone and its derivative ά-zearalenol. Appl Environ
Microbiol. 68:3545–3549. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Niderkorn V, Boudra H and Morgavi DP:
Binding of Fusarium mycotoxins by fermentative bacteria
in vitro. J Appl Microbiol. 101:849–856. 2006.
|
|
18.
|
Cheng B, Wan C, Yang S, Xu H, Wei H, Liu
J, et al: Detoxification of deoxynivalenol by Bacillus
strains. J Food Safety. 30:599–614. 2010.
|
|
19.
|
Lu Q, Liang X and Chen F: Detoxification
of zearalenone by viable and inactivated cells of Planococcus
sp. Food Control. 22:191–195. 2011. View Article : Google Scholar
|
|
20.
|
El-Nezami HS, Kankaanpaa PE, Salminen S
and Ahokas J: Ability of dairy strains of lactic acid bacteria to
bind a common food carcinogen, afatoxin B1. Food Chem Toxicol.
36:321–326. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Topcu A, Bulat T, Wishah R and Boyaci IH:
Detoxification of aflatoxin B1 and patulin by Enterococcus
faecium strains. Int J Food Microbiol. 139:202–205. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tuomola M, Vahva M and Kallio H:
High-performance liquid chromatography determination of skatole and
indole levels in pig serum, subcutaneous fat, and submaxillary
salivary glands. J Agr Food Chem. 44:1265–1270. 1996. View Article : Google Scholar
|
|
23.
|
Dreizen S and Spies TD: Further studies on
the association between the products of protein putrefaction and
dental caries activity. J Dent Res. 27:305–315. 1948. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Tittsler RP, Sandholzer LA and Callahan
ET: The bacteriostatic action of skatole on Gram-negative enteric
bacilli. J Infect Dis. 57:57–60. 1935. View Article : Google Scholar
|
|
25.
|
Li P, Tong L, Liu K and Wang YX:
Biodegradation of 3-methylindole by Pseudomonas putida LPC24
under oxygen limited conditions. Fresenius Environ Bull.
19:238–242. 2010.
|
|
26.
|
Squires EJ and Lundström K: Relationship
between cytochrome P4502E1 in liver and levels of skatole and its
metabolites in intact male pigs. J Anim Sci. 75:2506–2511.
1997.PubMed/NCBI
|
|
27.
|
Matal J, Matuskova Z, Tunkova A,
Anzenbacherova E and Anzenbacher P: Porcine CYP2A19, CYP2E1 and
CYP1A2 forms are responsible for skatole biotransformation in the
reconstituted system. Neuro Endocrinol Lett. 30:36–40.
2009.PubMed/NCBI
|
|
28.
|
Diaz GJ and Squires EJ: Metabolism of
3-methylindole by porcine liver microsomes: responsible cytochrome
P450 enzymes. Toxicol Sci. 55:284–292. 2000. View Article : Google Scholar : PubMed/NCBI
|