Introduction
Cardiovascular diseases (CVDs) are the leading cause
of death globally. Based on data from the World Health Organization
(WHO), an estimated 17.3 million people died from CVDs in 2008,
representing 30% of all global deaths (1). To date, the majority of patients with
CVDs have been treated with drug therapies. The main drugs on the
market are diuretics, vasodilators, anticoagulants, antiplatelet
agents and β-blockers. While these drugs have yielded desired
responses, they have also led to unwanted side-effects (2,3).
Multiherb therapy, as an essential component of traditional
medicine systems, has been utilized for thousands of years in China
and other countries. It has exhibited improved curative efficacies
and fewer side-effects, and has been used in the prevention of
disease (4). It has been revealed
that the majority of herbal medicines exhibit multiple
cardiovascular effects (5). Thus,
multiherb therapy may be one of the best conventional and
complementary medical approaches in the prevention and treatment of
CVDs.
Salvia miltiorrhiza (SM) and Panax
notoginseng (PN) have been widely used in combination in
Traditional Chinese Medicine (TCM) for the therapy of CVDs in China
and other countries, including the United States (6–8). It
has been demonstrated that these two herbs are compatible and have
a synergistic effect (7). However,
the molecular mechanisms underlying their compatibility have yet to
be clearly elucidated. Numerous computational pharmacological
studies, which have been generated using library analysis,
quantitative structure-activity relationship (QSAR),
receptor-ligand interaction and biological networks, have been
developed to clarify the pharmacology and efficacy of TCM (9,10).
Therefore, in the present study, we compared the computational
pharmacology of SM and PN at the molecular level, in order to
enhance the understanding of factors affecting compatibility in TCM
and to accelerate modern TCM development.
Materials and methods
Preparation of SM and PN chemical
databases
The structures identified in the medicinal herbs of
SM and PN were taken from the Chinese Herbal Drug Database and the
Handbook of the Constituents in Chinese Herb Original Plants
(11,12). The total number of compounds in SM
and PN was 53 and 57, respectively. These compounds were converted
into three-dimensional structures and energy optimizations were
performed using the Discovery Studio 2.0 (DS 2.0) software
(Accelrys Inc., San Diego, CA, USA), based on the Merck Molecular
Force Field (MMFF). Following this, the protocol of Cluster Ligands
was used to cluster the compounds from the SM and PN chemical
databases (13).
Calculation of molecular descriptors
The protocol from ‘Calculate Molecular Properties’
in the QSAR module of DS 2.0 was employed to calculate the
descriptors for the compounds from the SM and PN chemical
databases. The chemical space was constructed using 150 diversity
descriptors, including the molecular properties of one, two and
three dimensions (14,15). Principal component analysis (PCA)
was then performed to map the distribution of the compounds in
chemical space.
Molecular docking
The modern docking program LigandFit, within DS 2.0,
was used to perform the molecular docking. The crystal structures
of 16 key proteins associated with CVDs (16,17)
were downloaded from the Research Collaboratory for Structural
Bioinformatics (RCSB) protein data bank (PDB; Table I; www.rcsb.org). All
crystallographic water was removed from the file and hydrogen atoms
were added. The inhibitor from the PDB file was used to define the
active site. The compounds from the SM and PN chemical databases
were docked into the protein models. All docked structures were
sorted according to their DockScore. The compounds with the
top-five DockScores were selected as potential active compounds, as
described previously (18).
 | Table I.Sixteen proteins associated with
CVDs. |
Table I.
Sixteen proteins associated with
CVDs.
| Protein | PDB code | Protein | PDB code |
|---|
| TNF-α | 2AZ5 | Factor IXa | 1X7A |
| eNOS | 1M9J | Factor Xa | 1FJS |
| COX-1 | 1CQE | Factor VIIa | 1YGC |
| COX-2 | 6COX | Factor XI | 1ZSL |
| PPARγ | 2HFP | HMG-CoA | 1HW8 |
| HO-1 | 3TGM | ACE I | 1UZE |
| Thrombin | 1YPJ | ACE II | 1R4L |
| ER | 1X7J | Renin | 1BIL |
Network construction and analysis
Cytoscape 2.8.3 was used for network construction
(19). The potential active
compounds and their corresponding target proteins were connected to
each other to generate a drug-target (D-T) network. In this
network, the nodes represented compounds or proteins and the edges
represented the compound-target interactions. All data were
analyzed using Cytoscape plugins.
Results
Comparison of the SM and PN chemical
databases: Clustering distribution
The compounds from the SM and PN chemical databases
were clustered by employing the default settings of Cluster Ligands
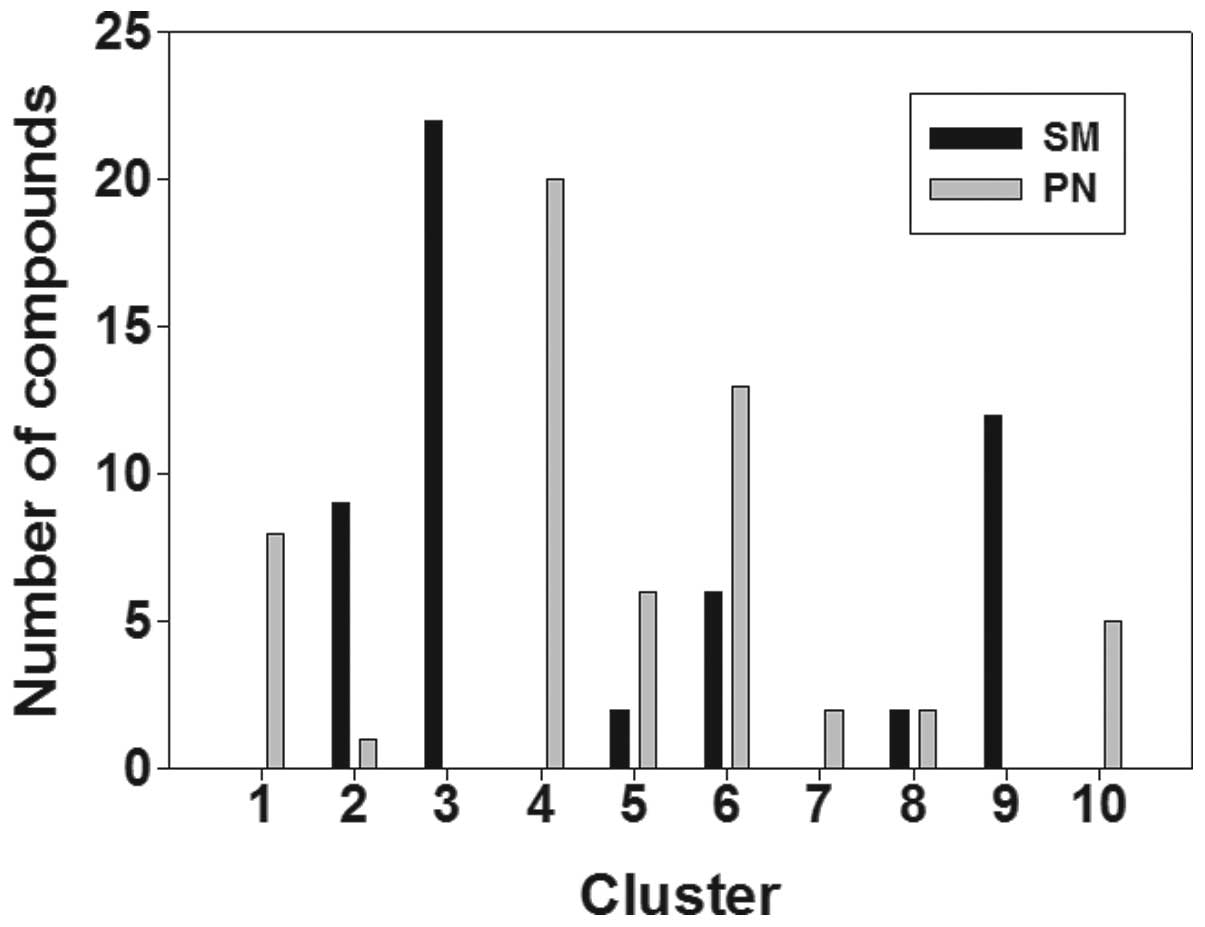
(Fig. 1). Fig. 1 shows that the compounds in SM were
attached to six clusters, known as clusters 2, 3, 5, 6, 8 and 9,
while the compounds in PN were attached to eight clusters, known as
clusters 1, 2, 4, 5, 6, 7, 8 and 10. These results indicate that SM
and PN have similarities and differences with regard to chemical
structure clustering.
Comparison of the SM and PN chemical
databases: Chemical space
Diversity descriptors (n=150) were used to map the
chemical space of the SM and PN chemical databases using PCA
(Fig. 2). A number of the key
molecular descriptors of the compounds from the two databases are
shown in Table II. The results
were as follows: i) Considerable dispersion was observed in the
first three principal components; ii) there was only a small
overlap between the databases of the two molecules in chemical
space; iii) the majority of the molecules did not violate
‘Lipinski’s rule of five’ (20).
 | Table II.Maximum, minimum and mean of the
molecular descriptors of the SM and PN chemical databases. |
Table II.
Maximum, minimum and mean of the
molecular descriptors of the SM and PN chemical databases.
| Descriptors | SM
| PN
|
|---|
| Maximum | Minimum | Mean | Maximum | Minimum | Mean |
|---|
| Molecular
weight | 718.61 | 154.12 | 326.40 | 1271.44 | 118.18 | 355.78 |
| No. of hydrogen
acceptors | 16 | 1 | 4.08 | 28 | 0 | 4.04 |
| No. of hydrogen
donors | 9 | 0 | 1.70 | 18 | 0 | 2.49 |
| AlogP | 8.08 | 0.61 | 3.57 | 10.41 | −4.54 | 4.34 |
| No. of rotatable
bonds | 14 | 0 | 1.98 | 19 | 0 | 7.96 |
| Molecular
volume | 438.35 | 88.49 | 219.33 | 853.72 | 86.43 | 270.14 |
| Molecular surface
area | 652.14 | 148.91 | 313.64 | 1221.90 | 128.43 | 378.57 |
| Molecular polar
surface area | 278.03 | 20.23 | 75.31 | 456.44 | 0 | 67.57 |
Comparison of the SM and PN chemical
databases: Compounds with potential biological activity
To investigate whether the compounds in SM and PN
were likely to be active in CVDs, the compounds with potential
biological activity were predicted using virtual docking. The
docking showed that various bioactive compounds in SM and PN
targeted multiple proteins associated with CVDs. The average number
of targets correlated with each compound in SM and PN were 5.0 and
3.6, respectively.
Comparison of the SM and PN chemical
databases: D-T network
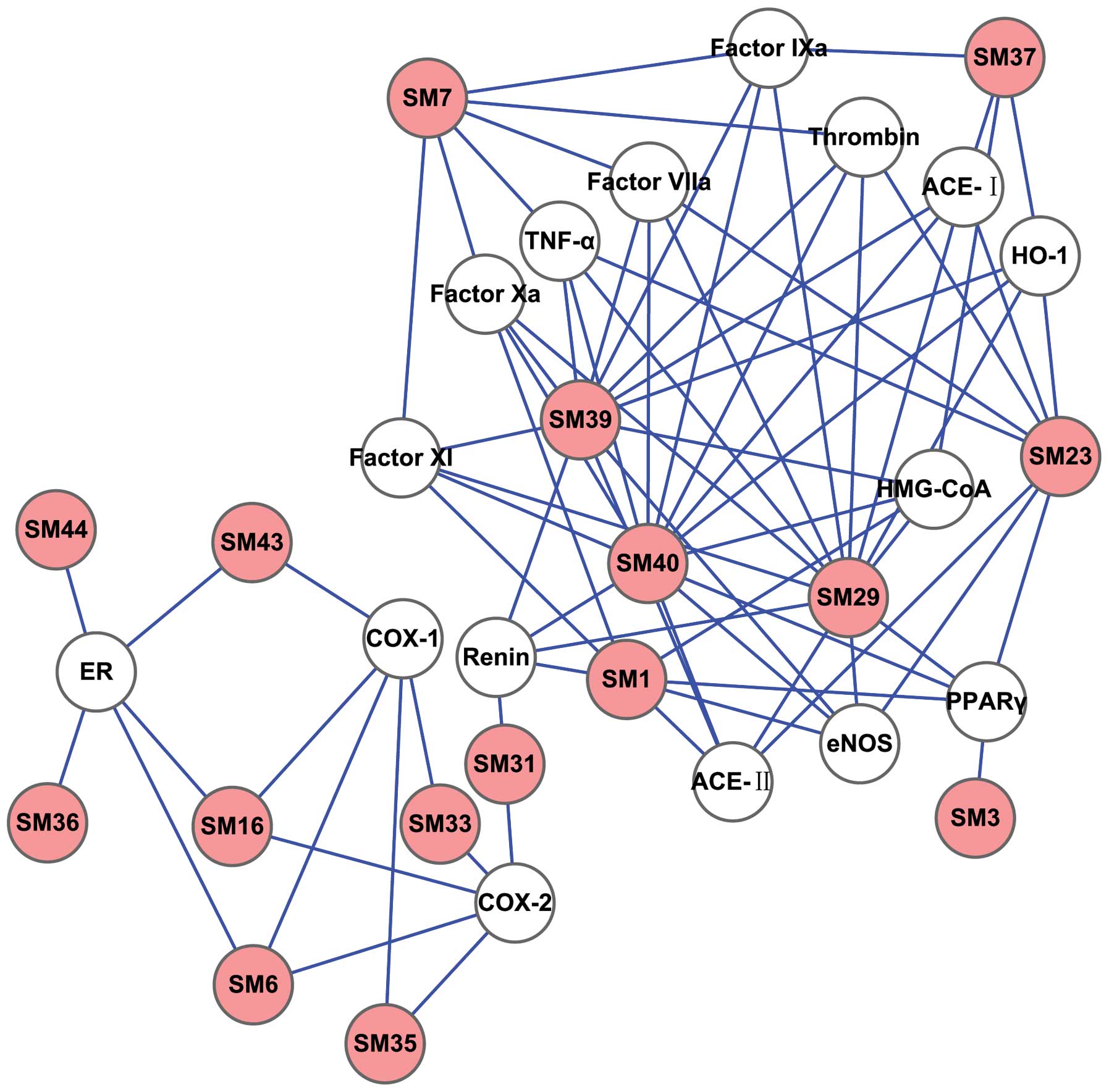
The D-T network was generated by connecting the
potential active compounds to their CVD-associated targets, in
order to further clarify the associations between the potentially
active compounds and their targets (Figs. 3 and 4). The simple parameters of the networks
for SM and PN are shown in Table
III. Plot parameters of these networks were used to
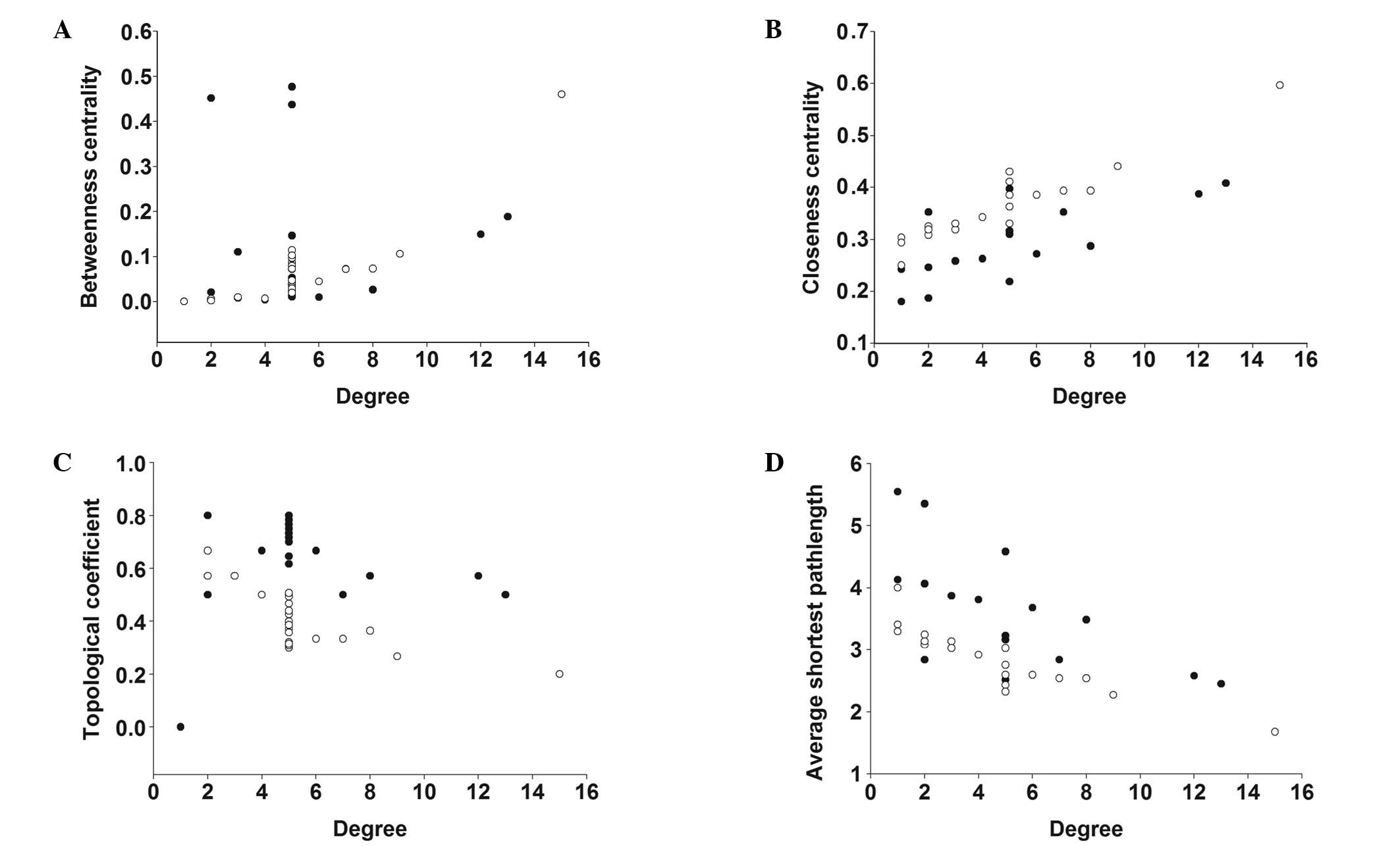
characterize the global map of D-T interactions (Fig. 5) and the degree of compound nodes
in the network was analyzed (Fig.
6). The key compounds with the top-five degrees in the two
networks are shown in Table IV.
These results demonstrated that SM and PN are able to act on
multiple targets and exhibit different modes of action between
compounds and targets.
 | Table III.Network properties of the SM and PN
D-T networks. |
Table III.
Network properties of the SM and PN
D-T networks.
| Parameters | SM D-T network | PN D-T network |
|---|
| Network
density | 0.161 | 0.114 |
| Network
heterogeneity | 0.596 | 0.652 |
| Network
centralization | 0.275 | 0.308 |
| Characteristic path
length | 3.544 | 2.762 |
| Average no. of
neighbors | 5.000 | 4.211 |
| Shortest paths | 992 (100%) | 1,406 (100%) |
 | Table IV.Key compounds with the top-five
degrees in the SM D-T network and PN D-T network. |
Table IV.
Key compounds with the top-five
degrees in the SM D-T network and PN D-T network.
SM D-T network
| PN D-T network
|
|---|
| Index | Chemical name | Degree | Index | Chemical name | Degree |
|---|
| SM29 | Monomethyl
lithospermate | 13 | PN50 | Quercetin | 15 |
| SM40 | Salvianolic acid
C | 13 | PN10 | Dicapryl
phthalate | 9 |
| SM39 | Salvianolic acid
A | 12 | PN11 | Diisocapryl
phthalate | 8 |
| SM23 | Methyl
rosmarinate | 8 | PN53 | Stigmasterol | 7 |
| SM1 | Baicalin | 7 | PN47 | Panaxynol | 6 |
Discussion
CVDs are the leading cause of mortality in the world
and pose a serious threat to human health. The diseases are complex
and multifactorial and are caused by environmental, genetic and
clinical risk factors (21).
Therefore, CVDs tend to result from multiple molecular
abnormalities. As a result, one drug acting on a single target may
not lead to the effective treatment of the CVDs (22,23),
and drug therapies addressing multiple targets have been receiving
increasing focus.
For many years, numerous preparations of the herb
pair comprising SM and PN have been used in the treatment of
patients with CVDs (24). In the
current study, a number of compounds in SM and PN, distributed
among the different class groups, are shown in Fig. 1. The results reflect the structural
diversity in the molecular composition of SM and PN and the
differences between them. SM and PN were shown to possess a broad
and different diversity in chemical space (Fig. 2), which indicates that they are
likely to exert different effects (15). The statistics of the drug-like
properties of the compounds from the SM and PN chemical databases
(Table II) revealed the mean
molecular weights to be 326.40 and 355.78, respectively; the mean
number of hydrogen bond acceptors was 4.08 and 4.04, respectively;
the mean number of hydrogen bond donors was 1.70 and 2.49,
respectively and the mean AlogP was 3.57 and 4.34, respectively.
According to the ‘rule of five’ (20), these compounds have desirable
drug-like properties that make them suitable for use as oral drugs
in humans. These results provided a good foundation for the
screening of suitable active compounds.
A docking screening protocol was used to identify
the compounds with multitarget potential for targets associated
with CVDs using LigandFit. The docking results showed that the
compounds in SM and PN exhibited potential biological activity with
one or more target proteins. Among these compounds, 81.25% of the
compounds in SM and 59.05% of the compounds in PN were able to act
on more than one target protein. To further compare the effects of
SM and PN on CVDs, we used the screening compounds and their
interaction targets to generate a bipartite graph of drug-target
interactions, in which a compound and a protein were connected to
each other if the protein was an action target of the compound,
giving rise to SM (Fig. 3) and PN
(Fig. 4) D-T networks. The
analyses of these networks (Fig.
5) showed minority common values of betweenness centrality,
closeness centrality, topological coefficients and shortest path
length, in addition to discriminating the detailed actions of SM
and PN on CVDs. Zhu et al (25) proposed that identifying the common
behavioral features from the network was likely to provide
important information to enable the understanding of the
drug-target interaction mechanisms in the human body; therefore,
the topological analysis may reflect global knowledge concerning
the particular properties of compounds and proteins involved in the
network. The results indicated that SM and PN exhibited different
modes of action. In addition, the compounds in SM and PN possessed
different degrees in the model of the D-T network (Fig. 6 and Table IV). The majority of the compounds
in Table IV have been described in
previous studies (26–29). As mentioned previously, the herb
pair consisting of SM and PN may possess a range of functions in
the treatment of CVDs, via different compounds combining with
different targets. Furthermore, the different modes of action of
the pair may result in SM and PN exerting synergistic effects in
CVDs. Zheng et al (7)
demonstrated that SM primarily acted to expand blood vessels, while
PN mainly participated in the protection of cardiac myocytes.
Therefore, the combination of these two herbs improves coronary
circulation and reduces the symptoms of myocardial ischemia. This
may further corroborate the proposal that SM and PN are able to
treat CVDs via different modes of action and that the combination
of the two herbs is able to enhance the therapeutic effects.
In conclusion, the results of the present study
demonstrated that: i) The compounds in SM and PN have diverse and
drug-like properties; ii) the compounds in SM and PN have
multitarget potential for targets associated with CVDs, and iii)
the combination of SM and PN may enhance their activities in
different potential multidrug combination therapies for CVDs.
Furthermore, the method of computational pharmacology is able to
intuitively trace out the different details of the structural
classification, chemical space and modes of action of the compounds
in SM and PN. This may be used as a new method for the
identification of SM and PN, and to enable the improved
understanding of herb pairs at the molecular level.
Abbreviations:
|
TCM
|
Traditional Chinese Medicine;
|
|
SM
|
Salvia miltiorrhiza;
|
|
PN
|
Panax notoginseng ;
|
|
CVDs
|
cardiovascular diseases;
|
|
TNF-α
|
tumor necrosis factor-α;
|
|
eNOS
|
endothelial nitric oxide synthase;
|
|
COX
|
cyclooxygenase;
|
|
HO
|
heme oxygenase;
|
|
PPARγ
|
peroxisome proliferator activated
receptor γ;
|
|
ER
|
estrogen receptor;
|
|
HMG-CoA
|
3-hydroxy-3-methylglutaryl coenzyme
A;
|
|
ACE
|
angiotensin-converting enzyme;
|
|
D-T network
|
drug-target network
|
Acknowledgements
This study was supported by the
Developmental Fund of Chen Keji Integrative Medicine
(CKJ2010032).
References
|
1.
|
Alwan A, Armstrong T, Bettcher D, Branca
F, Chisholm D, Ezzati M, Garfield R, MacLean D, Mathers C, Mendis
S, Poznyak V, Riley L, Tang KC and Wild C: Global status report on
noncommunicable diseases 2010. WHO Library
Cataloguingin-Publication Data; Geneva: pp. 100–143. 2011
|
|
2.
|
Li SB: Adverse drug reaction and
prevention of cardiovascular drugs. Hainan Med J. 22(3): 14–19.
2011.(In Chinese).
|
|
3.
|
Toyoshima H, Takahashi K and Akera T: The
impact of side effects on hypertension management: a Japanese
survey. Clin Ther. 19:1458–1469. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Wang S, Hu Y, Tan W, Wu X, Chen R, Cao J,
Chen M and Wang Y: Compatibility art of traditional Chinese
medicine: from the perspective of herb pairs. J Ethnopharmacol.
143:412–423. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Frishman WH, Sinatra ST and Moizuddin M:
The use of herbs for treating cardiovascular disease. Semin Integr
Med. 2:23–35. 2004. View Article : Google Scholar
|
|
6.
|
Liang CZ: Focus on the theory of
precaution of disease in Tradition Chinese Medicine from the
function of compound Danshen dripping pill to the heart and blood
vessel’s illness. Chin Arch Trad Chin Med. 26:643–644. 2008.(In
Chinese).
|
|
7.
|
Zheng Q, Peng CC, Shen ML and Yang M:
Study on compatibility of Radix et Rhizoma Salviae
miltiorrhizae and Radix et Rhizoma notoginseng. Chin J Exp Trad
Med Formulae. 15(2): 83–86. 2009.(In Chinese).
|
|
8.
|
Cheng TO: Cardiovascular effects of
Danshen. Int J Cardiol. 121:9–22. 2007. View Article : Google Scholar
|
|
9.
|
Zheng CS, Xu XJ, Liu XX and Ye HZ:
Computational pharmacology of Jingzhi Tougu Xiaotong granule in
preventing and treating osteoarthritis. Acta Phys Chim Sin.
26:775–783. 2010.(In Chinese).
|
|
10.
|
Ma S, Feng C, Zhang X, Dai G, Li C, Cheng
X, Liu P, Ju W and Yu H: The multi-target capabilities of the
compounds in a TCM used to treat sepsis and their in silico
pharmacology. Complement Ther Med. 21:35–41. 2013.PubMed/NCBI
|
|
11.
|
Qiao X, Hou T, Zhang W, Guo S and Xu X: A
3D structure database of components from Chinese traditional
medicinal herbs. J Chem Inf Comput Sci. 42:481–489. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Zhou JX, Xie GR and Yang XD: Handbook of
the Constituents in Chinese Herb Original Plants. 1st edition.
Chemical Industry Press; Beijing: pp. 1165–1211. 2004
|
|
13.
|
Hassan M, Bielawski JP, Hempel JC and
Waldman M: Optimization and visualization of molecular diversity
and combinatorial libraries. Mol Divers. 2:64–74. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Zheng CS, Xu XJ, Ye HZ, Wu GW, Li XH,
Huang SP and Liu XX: Computational approaches for exploring the
potential synergy and polypharmacology of Duhuo Jisheng Decoction
in the therapy of osteoarthritis. Mol Med Rep. 7:1812–1818.
2013.PubMed/NCBI
|
|
15.
|
Dobson CM: Chemical space and biology.
Nature. 432:824–828. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wu DH and Xu XJ: Computational simulation
of benefiting Qi and activating blood mechanism of Traditional
Chinese Medicine. Acta Phys Chim Sin. 25:446–450. 2009.(In
Chinese).
|
|
17.
|
Gu JY, Yuan G, Zhu YH and Xu XJ:
Computational pharmacological studies on cardiovascular disease by
Qishen Yiqi Diwan. Sci China B. 52:1871–1878. 2009. View Article : Google Scholar
|
|
18.
|
Zheng CS, Ye HZ, Cai LL, Chen JS, Wei LS
and Liu XX: Discussion on multi-component and multi-target pattern
of Liuwei Dihuang pill in the treatment of osteoarthritis on the
basis of computer simulation. J Trad Chin Orthop Traumatol.
25:11–13. 182013.(In Chinese).
|
|
19.
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Cooney MT, Dudina A, D’Agostino R and
Graham IM: Cardiovascular risk-estimation systems in primary
prevention: do they differ? Do they make a difference? Can we see
the future? Circulation. 122:300–310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Baur JA and Sinclair DA: Therapeutic
potential of resveratrol: the in vivo evidence. Nat Rev Drug
Discov. 5:493–506. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Hopkins AL: Network pharmacology: the next
paradigm in drug discovery. Nat Chem Biol. 4:682–690. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Chu Y, Zhang L, Wang XY, Guo JH, Guo ZX
and Ma XH: The effect of Compound Danshen Dripping Pills, a Chinese
herb medicine, on the pharmacokinetics and pharmacodynamics of
warfarin in rats. J Ethnopharmacol. 137:1457–1461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zhu M, Gao L, Li X, Liu Z, Xu C, Yan Y,
Walker E, Jiang W, Su B, Chen X and Lin H: The analysis of the
drug-targets based on the topological properties in the human
protein-protein interaction network. J Drug Target. 17:524–532.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Perez-Vizcaino F and Duarte J: Flavonols
and cardiovascular disease. Mol Aspects Med. 31:478–494. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ho JH and Hong CY: Salvianolic acid: small
compounds with multiple mechanisms for cardiovascular protection. J
Biomed Sci. 18:302011. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Ji XF and Shi DH: Advances in studies of
baicalin on cardiovascular and cerebrovascular pharmacology. Strait
Pharm J. 18:10–12. 2006.(In Chinese).
|
|
29.
|
Jiang LP, Nie BM, Lu HM, Gan LJ, Chen HZ
and Lu Y: Inhibitory effect of panaxynol on the proliferation of
rat aortic smooth muscle cell and its mechanisms. Chin Pharmacol
Bull. 21:1313–1319. 2005.(In Chinese).
|