Identification of genes in HepG2 cells that respond to DNA methylation and histone deacetylation inhibitor treatment
- Authors:
- Published online on: June 18, 2014 https://doi.org/10.3892/etm.2014.1789
- Pages: 813-817
Abstract
Introduction
Hepatic carcinoma is the sixth most common cancer worldwide and the third most common cause of mortality from cancer with 626,000 cases and 598,000 mortalities annually (1). In China, there are 360,000 cases of hepatic carcinoma and 350,000 associated mortalities a year (2), and hepatic carcinoma is the second most common cause of cancer-associated mortalities (1,3). Hepatitis B virus and aflatoxins are considered the major and common factors attributed to the etiology of liver cancer, and they can act individually or synergistically on the liver to cause cancer (4,5). Other factors, including hepatitis C virus, genetic susceptibility or genetic polymorphisms, may also have an important role in the etiology of liver cancer (6).
Previous studies have investigated the mechanism of hepato-carcinogenesis (7,8). The majority of these studies have focused on the genetic changes in key tumor suppressor genes and oncogenes; however, it has been suggested that epigenetic disruption of gene expression may also have an important role in the development of cancer (9). Epigenetic events have been found to be involved in the etiology of a wide variety of types of human cancer, including hepatic carcinoma. The current definition of epigenetics is the study of heritable changes in gene expression that occur independently from changes in the primary DNA sequence (10). The heritability of gene expression patterns is primarily mediated by epigenetic modifications, which include DNA methylation, chromatin remodeling, histone replacement and alterations to histone tails (8,11,12). DNA methylation is the most extensively studied epigenetic modification in mammals, and it provides a stable gene silencing mechanism that has an important role in the regulation of gene expression and chromatin architecture (10). Several studies have reported that there are somatically acquired DNA methylation changes in various tumor-suppressor genes and other cancer-associated genes (13,14). Histone deacetylation is a type of histone modification that may regulate key cellular processes, including transcription, DNA replication and DNA repair (15). DNA methylation and histone deacetylation may work independently or in concert to alter gene expression during tumorigenesis. Therefore, in the present study, the effect of inhibiting DNA methylation and histone deacetylation in HepG2 cells was investigated to determine the potential role of epigenetic modifications in the development and treatment of hepatic carcinoma, and to explore a novel therapeutic strategy for the treatment of hepatic carcinoma.
Materials and methods
Research materials and gene chip
In order to explore the effect of DNA methylation and histone deacetylation on hepatoma cells, the HepG2 cell line was used. The cells had been treated with 5-aza-2′-deoxycytidine (5-aza-dC; aza), trichostatin A (TSA), and a combination of aza and TSA to inhibit DNA methylation, histone deacetylation and both methylation and deacetylation, respectively. The gene expression profiles of the treated cells were compared with those of the control group to investigate the effects of methylation and deacetylation on liver cancer cells. GSE5230 sample data from the Gene Expression Omnibus (GEO) database was used (16), which included 4 gene chips of the treatment by aza, TSA, combination of aza and TSA and the control group, respectively.
Acquisition of the differentially expressed genes
The samples were identified and the microarray data were analyzed using the R software (v.2.13.0) (17) platform, as well as GEOquery (18) and the limma package to further process the data. GEOquery obtains chip expression profiling data from the GEO database quickly, whilst limma can be used to statistically analyze the differentially expressed genes (19,20). The GEOquery package was used to obtain data of chip expression profiling that had already been preprocessed, and the chip data as transformed with log2. The expression profiles of the HepG2 cells treated with aza, TSA, aza and TSA and the control group were then compared, and the differentially expressed genes inhibited by methylation and acetylation were analyzed using the linear regression model package limma.
Gene Otology (GO) analysis of the differentially expressed genes
In order to investigate the changes in the differentially expressed genes at the cellular level and their functional clustering, classification of gene function and position was performed using GO (21), using the GOEAST platform (22). In the present study, a hyper-geometric algorithm was selected for the statistical analysis. The entire microarray probe was used as a background control and the differentially expressed genes from biological processes were clustered; thus, the effect of these differentially expressed genes on the cells was determined.
Biological pathway data
In order to investigate the changes induced in the cells as a result of the inhibition of DNA methylation and histone deacetylation at the molecular level, the effects of these modifications on biological pathways were examined. All metabolic and non-metabolic pathways were acquired from the current public open access database WikiPathways(http://www.wikipathways.org) (23,24), and the WikiPathways clustering analysis of differentially expressed genes was achieved through the Gene Set Analysis Toolkit V2 platform (25,26), in order to determine the changes in the signal pathways of HepG2 cells.
Identification of potential target sites for regulatory transcription factors
Based on the gene annotation data arranged by the MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) database, analyzed by gene abundance, with statistical calculations conducted with a hypergeometric algorithm and calibrated by Benjamini and Hochberg (BH) procedure, the potential target sites for regulation by transcription factors were obtained.
Results
Identification of differentially expressed genes following the inhibition of methylation and deacetylation
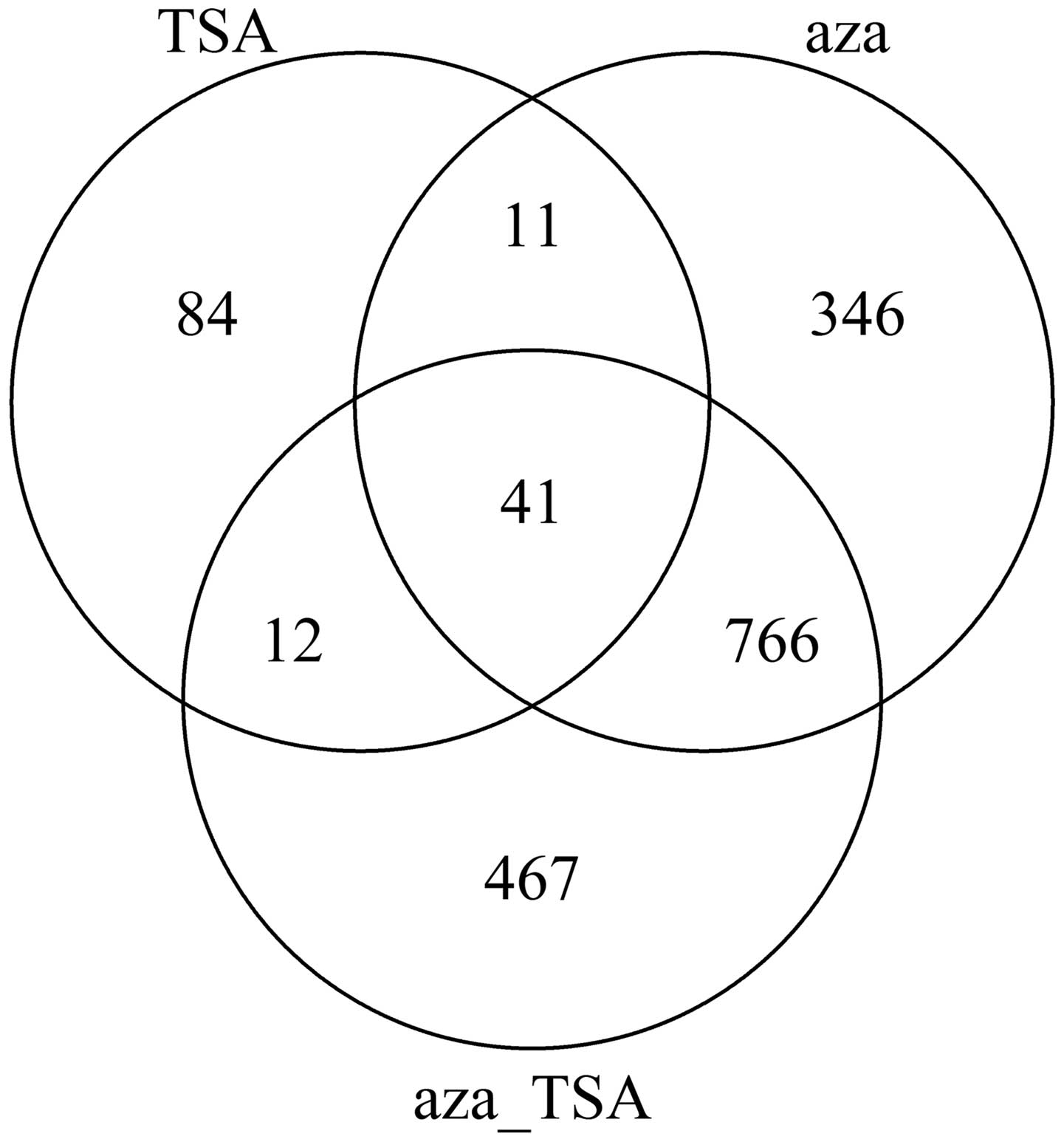
The data were analyzed using a t-test (20) modified by Bayesian model in order to obtain the differentially expressed genes. P-values were obtained for all the genes, and they were corrected as the false discovery rate (FDR). P<0.05 was considered to indicate a statistically significant difference. The numbers of genes with changes in expression levels are shown in Fig. 1.
As seen in Fig. 1, treatment with aza or TSA induced numerous changes in gene expression in HepG2 cells. The number of the altered genes following aza treatment was larger compared with that following TSA treatment. The results indicate that inhibition of DNA methylation and histone acetylation affects gene expression of HepG2 cells; however, methylation has a more significant contribution to the gene expression and regulation of liver cancer cells.
Biological pathway enrichment regulated by DNA methylation and histone acetylation
Since the inhibition of DNA methylation and histone deacetylation caused changes in the expression in certain genes, changes in the biological pathways of hepatoma cells following the inhibition of DNA methylation and histone deacetylation was further investigated. The differentially expressed genes were selected and WikiPathways sub-pathway enrichment analysis was performed. The genes were clustered by hypergeometric algorithm and then multiplex detection was proofread using the BH algorithm in order to identify changes in the signaling pathways of hepatoma cells. Biological pathways significantly changed under the limiting conditions (corrected to P<0.1) with at least two genes in the signaling pathway are shown in Table I.
Table IChanges in biological pathways following inhibition of DNA methylation and histone acetylation by aza and TSA. |
As the expression of only a few genes changed following treatment with TSA, clustering of only one signaling pathway, the transforming growth factor (TGF)-β signaling pathway, was observed. The TGF-β signaling pathway has very important roles in the body, including during embryonic development, cell growth, differentiation and apoptosis, as well as in intracellular metabolic balance. Therefore, histone deacetylation appears to have a critical effect on liver cancer cells.
DNA methylation was inhibited following treatment with aza, resulting in a series of genes being expressed differentially. Multiple biological pathways are associated with these differentially expressed genes, including signal transduction-related integrin-mediated cell adhesion, the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway, the α6β4 signaling pathway, prostaglandin synthesis and regulation, the prolactin signaling pathway, metabolism-associated fatty acid β-oxidation, fluoropyrimidine activity, the drug-related irinotecan pathway and the cell motility-associated complement and coagulation cascades pathway.
The signaling pathways altered following treatment with Aza and TSA were broadly similar to those altered following treatment with aza alone, which include striated muscle contraction, the irinotecan pathway, AMPK signaling, the α6β4 signaling pathway, fluoropyrimidine activity and fatty acid β-oxidation. Furthermore, Aza and TSA co-treatment had a significant influence on fatty acid metabolism in HepG2 cells; however, mitochondrial long chain-fatty acid β-oxidation and fatty acid biosynthesis were indicated to be unaffected, with the exception of fatty acid β-oxidation.
GO clustering of the differentially expressed genes
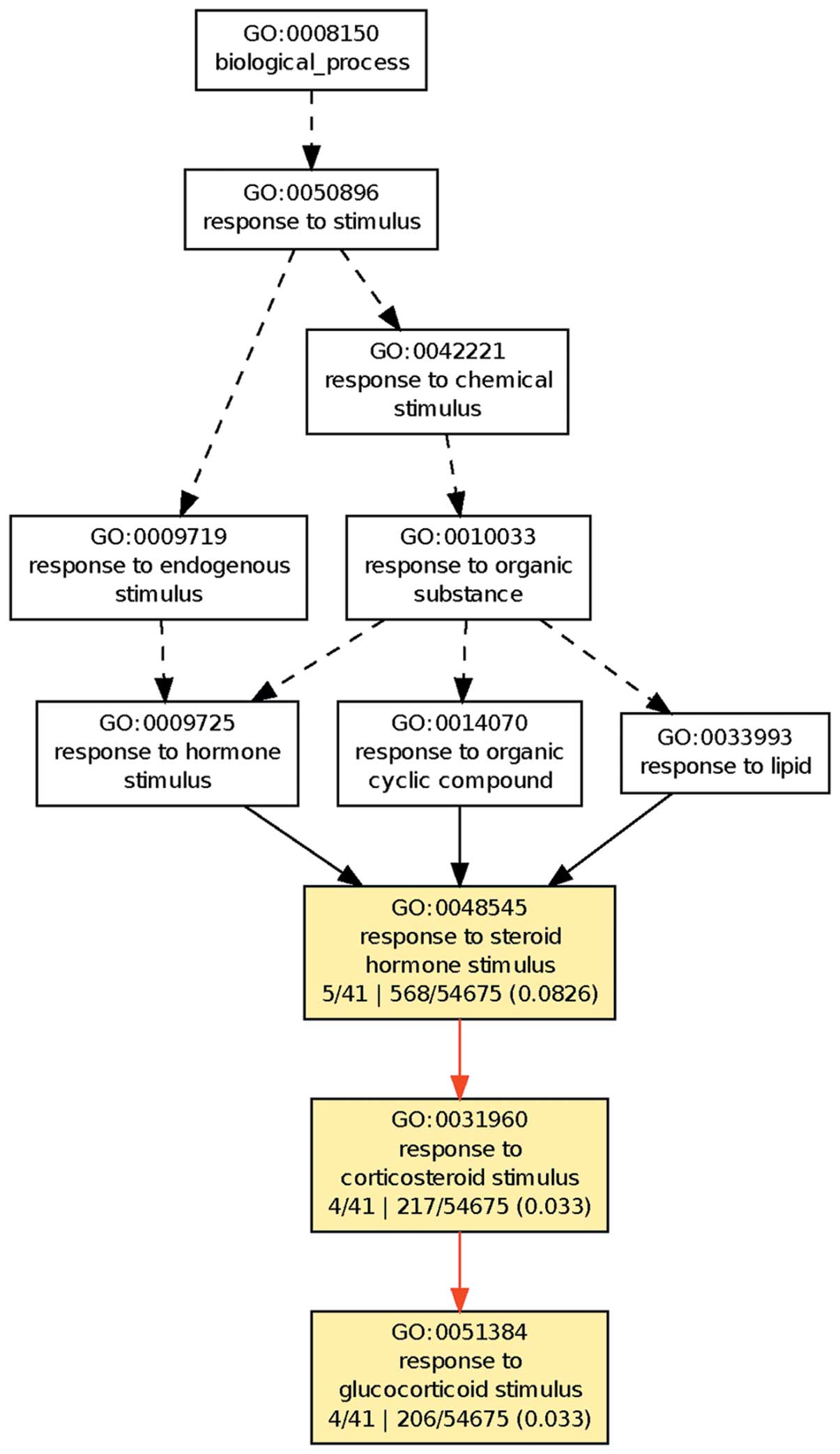
In the organism, various means are required for regulation of the more important physiological processes. Therefore, the present study focused on the 41 genes expressed differentially for all three treatments. GO clustering was performed on their physiological processes using the GOEAST platform, as shown in Fig. 2.
The results demonstrated that these 41 genes clustered on the cell response to steroid hormones, in particular glucocorticoids. This suggests that DNA methylation and histone deacetylation have important roles in the regulation of the response to glucocorticoid in hepatoma cells.
Analysis of target sites of the potential transcription factors
The spatial structure of the chromosome is altered as a result of the epigenetic modifications of DNA and histones, and this then alters the binding ability of trans-regulatory elements. As an important class of trans-regulatory elements, transcription factors may be a cause of the changes of gene expression following the inhibition of DNA methylation or histone deacetylation. Therefore, in the present study common binding sites for transcription factors shared by the differentially expressed genes were identified. Upstream sequences of the differentially expressed genes were used to investigate the potential target sites for the transcription factors, and the hypergeometric clustering algorithm was used, with proofreading of the P-value with the BH algorithm, and 10 target sites for the transcription factors were then identified (Table II).
Discussion
The results from the WikiPathways clustering analysis demonstrated that inhibition of histone acetylation in HepG2 cells by exposure to TSA significantly affected the TGF-β signaling pathway, which indicates that histone deacetylation has an important role in the TGF-β signaling pathway in HepG2 cells. Furthermore, among the significantly upregulated genes in the TGF-β signaling pathway, lymphoid-enhancing factor 1 (LEF1) was of particular interest, as it is known to have a functional role in the Wnt/β-catenin pathway, another important pathway for tumor growth and invasion (27). Inhibition of histone acetylation in HepG2 cells may downregulate LEF1 expression, inhibiting the growth of HepG2 cells. In addition, bone morphogenetic protein 4 (BMP4) was found to be upregulated following inhibition of histone deacetylation. BMP4 is a member of the TGF-β family and is found in the liver. Previous studies have shown that BMP4 is constitutively expressed in the peribiliary stroma and endothelial cells in the liver and that its expression is downregulated following hepatectomy (28,29); in addition, BMP4 serves as an antiproliferative factor in hepatocyte proliferation (28).
Altering the TGF-β signaling pathway as a possible therapeutic treatment for cancer has been previously investigated in numerous studies (30,31). Therefore, the inhibition of histone acetylation in HepG2 cells to alter the TGF-β signaling pathway may provide targeted therapy for hepatic carcinoma; however, further investigation is required to determine the detailed mechanisms of TGF-β production and activation.
Treatment with aza and the combination of aza and TSA inhibited DNA methylation in HepG2 cells, which resulted in alterations in intracellular biological pathways, including integrin-mediated cell adhesion. Several of the signaling transduction pathways have important roles in growth, metabolism and regulation of differentiation in HepG2 cells. For example, TNFSF13, a member of the tumor necrosis factor family, was found to be upregulated following treatment with aza. TNFSF13 has a pathogenic role in the microenvironments of solid and hematological tumors (32). Elevated serum levels of TNFSF13 have been reported in oral cavity cancers (33), and are correlated with increased serum TNF levels, angiogenesis and poor prognosis in multiple myeloma (34). In addition, the inhibition of DNA methylation following treatment with aza and TSA resulted in changes in the AMPK signaling pathway. AMPK is a master regulator of energy homeostasis and is involved in the regulation of a number of physiological processes, including the β-oxidation of fatty acids, lipogenesis and protein and cholesterol synthesis. Previous studies have demonstrated that changes in these processes occur during cancer due to alterations in AMPK activity within cancer cells or in their periphery (35). The results of the present study demonstrated that tumor suppressor proteins tuberous sclerosis complex (TSC) 1 and 2, which are substrates of AMPK, were differentially expressed and tumor suppressor p53 was upregulated. In addition, DNA methylation has been investigated as a potential biomarker and therapeutic target in malignant tumors (36). Furthermore, inhibition of DNA methylation has a similar therapeutic effect as irinotecan, which is an effective drug for the treatment of certain types of cancer, including intestinal cancer and small cell carcinoma (37). Therefore, inhibition of DNA methylation and histone acetylation may provide a novel therapeutic treatment for hepatic carcinoma.
In addition, the clustering of the GO physiological processes of the 41 differentially expressed genes for the three treatment groups in the present study showed that the response of the HepG2 cells to glucocorticoids changed. Glucocorticoids are a class of steroid hormones secreted by zona fasciculata in the adrenal cortex, and have a role in the regulation of glucose and fat metabolism, and protein biosynthesis and metabolism (38,39). In addition, they may inhibit the immune response and have anti-inflammatory effects (38,40). Therefore, inhibition of DNA methylation and histone acetylation not only affects the metabolism of HepG2 cells, but also the immune activity.
Furthermore, a large number of the genes that were found to be differentially expressed following inhibition of DNA methylation and histone deacetylation may have the same target sites for transcription factors, and these sites may have an important role in the regulation of gene expression. For example, the differentially expressed gene BMP4 mentioned previously may be regulated by SMAD1. BMP4 signal transduction is dependent on SMAD phosphorylation via alk3 and SMAD signaling is associated with decreased hepatocyte proliferation following hepatectomy (41).
In conclusion, the present study identified a range of differentially expressed genes associated with DNA methylation and histone deacetylation blockage in HepG2 cells. Further studies of these genes and their regulation may aid in elucidating the underlying mechanism of the development of hepatocellular carcinoma.
References
|
Jemal A, Bray F, Center MM, Ferlay J, Ward E and Forman D: Global cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar | |
|
Chen JG, Zhang SW and Chen WQ: Analysis of liver cancer mortality in the national retrospective sampling survey of death causes in China, 2004–2005. Zhonghua Yu Fang Yi Xue Za Zhi. 44:383–389. 2010.(In Chinese). | |
|
Chen JG and Zhang SW: Liver cancer epidemic in China: past, present and future. Semin Cancer Biol. 21:59–69. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Luo RH, Zhao ZX, Zhou XY, Gao ZL and Yao JL: Risk factors for primary liver carcinoma in Chinese population. World J Gastroenterol. 11:4431–4434. 2005.PubMed/NCBI | |
|
Groopman JD, Johnson D and Kensler TW: Aflatoxin and hepatitis B virus biomarkers: a paradigm for complex environmental exposures and cancer risk. Cancer Biomark. 1:5–14. 2005.PubMed/NCBI | |
|
Chen TH, Chen CJ, Yen MF, et al: Ultrasound screening and risk factors for death from hepatocellular carcinoma in a high risk group in Taiwan. Int J Cancer. 98:257–261. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Aguirre-Ghiso JA: Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 7:834–846. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ducasse M and Brown MA: Epigenetic aberrations and cancer. Mol Cancer. 5:602006. View Article : Google Scholar | |
|
Dolinoy DC, Weidman JR and Jirtle RL: Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 23:297–307. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Esteller M: Epigenetics in cancer. N Engl J Med. 358:1148–1159. 2008. View Article : Google Scholar | |
|
Jenuwein T: The epigenetic magic of histone lysine methylation. FEBS J. 273:3121–3135. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Barkess G: Chromatin remodeling and genome stability. Genome Biol. 7:3192006. View Article : Google Scholar : PubMed/NCBI | |
|
Esteller M: Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 8:286–298. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Vucic EA, Brown CJ and Lam WL: Epigenetics of cancer progression. Pharmacogenomics. 9:215–234. 2008. View Article : Google Scholar | |
|
Kouzarides T: Chromatin modifications and their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Dannenberg LO and Edenberg HJ: Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 7:1812006. View Article : Google Scholar : PubMed/NCBI | |
|
R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008 | |
|
Davis S and Meltzer PS: GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Diboun I, Wernisch L, Orengo C and Koltzenburg M: Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI | |
|
Smyth GK: Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3:2004.PubMed/NCBI | |
|
Ashburner M, Ball CA, Blake JA, et al: Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng Q and Wang XJ: GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36:W358–W363. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Kelder T, van Iersel MP, Hanspers K, et al: WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 40:D1301–D1307. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR and Evelo C: WikiPathways: pathway editing for the people. PLoS Biol. 6:e1842008. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang B, Kirov S and Snoddy J: WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Duncan D, Prodduturi N and Zhang B: WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 11(Suppl 4): P102010. View Article : Google Scholar | |
|
Jeanes A, Gottardi CJ and Yap AS: Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 27:6920–6929. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Do N, Zhao R, Ray K, et al: BMP4 is a novel paracrine inhibitor of liver regeneration. Am J Physiol Gastrointest Liver Physiol. 303:G1220–G1227. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Xia Y, Babitt JL, Sidis Y, Chung RT and Lin HY: Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 111:5195–5204. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Giannelli G, Mazzocca A, Fransvea E, Lahn M and Antonaci S: Inhibiting TGF-β signaling in hepatocellular carcinoma. Biochim Biophys Acta. 1815:214–223. 2011. | |
|
Ikushima H and Miyazono K: TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 10:415–424. 2010. | |
|
Mackay F, Sierro F, Grey ST and Gordon TP: The BAFF/APRIL system: an important player in systemic rheumatic diseases. Curr Dir Autoimmun. 8:243–265. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Jablonska E, Slodczyk B, Wawrusiewicz-Kurylonek N, et al: Overexpression of B cell-activating factor (BAFF) in neutrophils of oral cavity cancer patients-preliminary study. Neoplasma. 58:211–216. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Fragioudaki M, Tsirakis G, Pappa CA, et al: Serum BAFF levels are related to angiogenesis and prognosis in patients with multiple myeloma. Leuk Res. 36:1004–1008. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Brown KA, Samarajeewa NU and Simpson ER: Endocrine-related cancers and the role of AMPK. Mol Cell Endocrinol. 366:170–179. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Akhurst RJ and Derynck R: TGF-beta signaling in cancer - a double-edged sword. Trends Cell Biol. 11:S44–S51. 2001.PubMed/NCBI | |
|
de Jong FA, de Jonge MJ, Verweij J and Mathijssen RH: Role of pharmacogenetics in irinotecan therapy. Cancer Lett. 234:90–106. 2006.PubMed/NCBI | |
|
Taves MD, Gomez-Sanchez CE and Soma KK: Extra-adrenal glucocorticoids and mineralocorticoids: evidence for local synthesis, regulation, and function. Am J Physiol Endocrinol Metab. 301:E11–E24. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Rose AJ, Vegiopoulos A and Herzig S: Role of glucocorticoids and the glucocorticoid receptor in metabolism: insights from genetic manipulations. J Steroid Biochem Mol Biol. 122:10–20. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Amsterdam A, Tajima K and Sasson R: Cell-specific regulation of apoptosis by glucocorticoids: implication to their anti-inflammatory action. Biochem Pharmacol. 64:843–850. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Goldman DC, Bailey AS, Pfaffle DL, Al Masri A, Christian JL and Fleming WH: BMP4 regulates the hematopoietic stem cell niche. Blood. 114:4393–4401. 2009. View Article : Google Scholar : PubMed/NCBI |