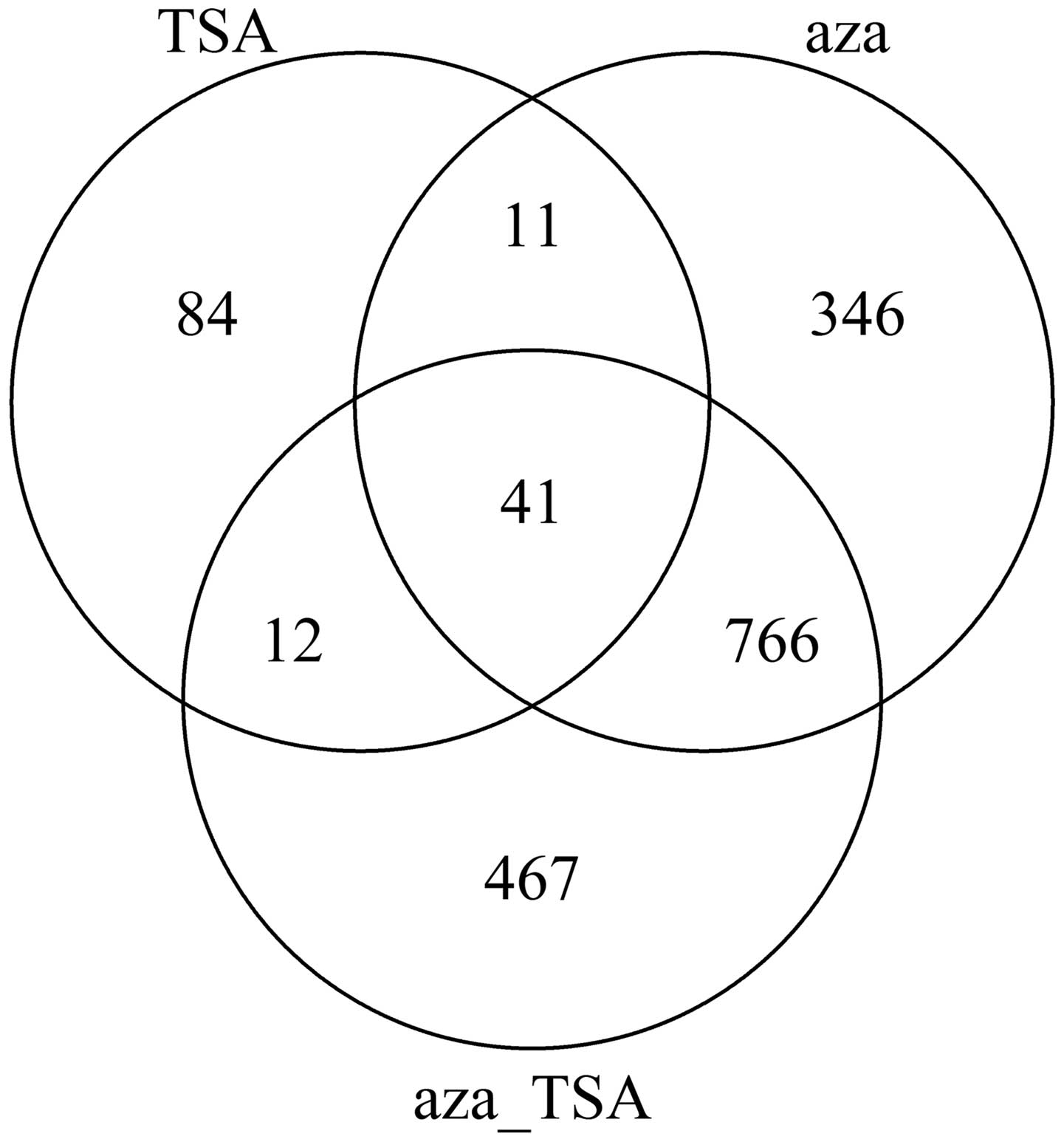
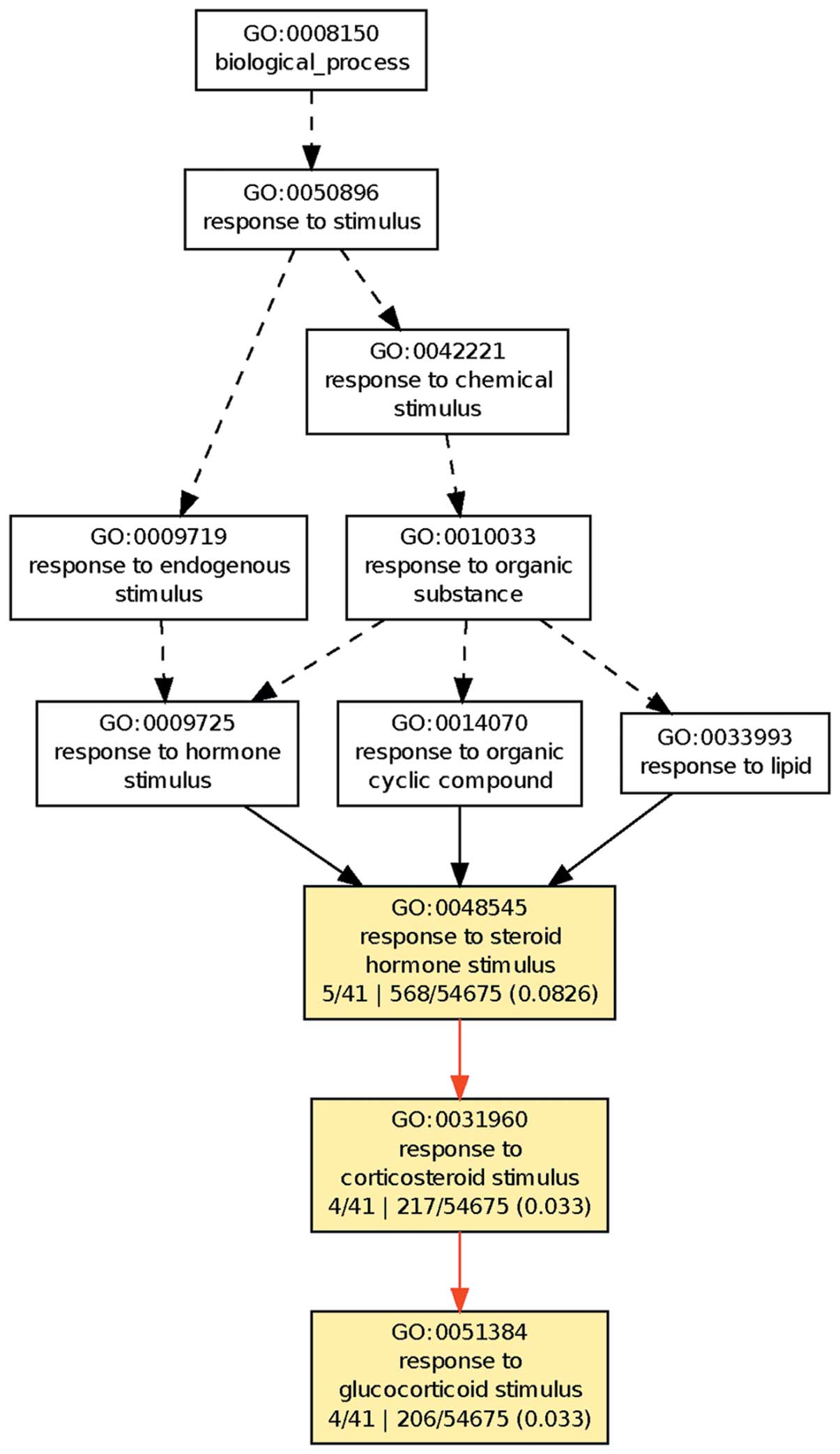
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Chen JG, Zhang SW and Chen WQ: Analysis of
liver cancer mortality in the national retrospective sampling
survey of death causes in China, 2004–2005. Zhonghua Yu Fang Yi Xue
Za Zhi. 44:383–389. 2010.(In Chinese).
|
|
3
|
Chen JG and Zhang SW: Liver cancer
epidemic in China: past, present and future. Semin Cancer Biol.
21:59–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo RH, Zhao ZX, Zhou XY, Gao ZL and Yao
JL: Risk factors for primary liver carcinoma in Chinese population.
World J Gastroenterol. 11:4431–4434. 2005.PubMed/NCBI
|
|
5
|
Groopman JD, Johnson D and Kensler TW:
Aflatoxin and hepatitis B virus biomarkers: a paradigm for complex
environmental exposures and cancer risk. Cancer Biomark. 1:5–14.
2005.PubMed/NCBI
|
|
6
|
Chen TH, Chen CJ, Yen MF, et al:
Ultrasound screening and risk factors for death from hepatocellular
carcinoma in a high risk group in Taiwan. Int J Cancer. 98:257–261.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aguirre-Ghiso JA: Models, mechanisms and
clinical evidence for cancer dormancy. Nat Rev Cancer. 7:834–846.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ducasse M and Brown MA: Epigenetic
aberrations and cancer. Mol Cancer. 5:602006. View Article : Google Scholar
|
|
9
|
Dolinoy DC, Weidman JR and Jirtle RL:
Epigenetic gene regulation: linking early developmental environment
to adult disease. Reprod Toxicol. 23:297–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar
|
|
11
|
Jenuwein T: The epigenetic magic of
histone lysine methylation. FEBS J. 273:3121–3135. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barkess G: Chromatin remodeling and genome
stability. Genome Biol. 7:3192006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vucic EA, Brown CJ and Lam WL: Epigenetics
of cancer progression. Pharmacogenomics. 9:215–234. 2008.
View Article : Google Scholar
|
|
15
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dannenberg LO and Edenberg HJ: Epigenetics
of gene expression in human hepatoma cells: expression profiling
the response to inhibition of DNA methylation and histone
deacetylation. BMC Genomics. 7:1812006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
R Development Core Team. R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2008
|
|
18
|
Davis S and Meltzer PS: GEOquery: a bridge
between the Gene Expression Omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Diboun I, Wernisch L, Orengo C and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smyth GK: Linear models and empirical
Bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:2004.PubMed/NCBI
|
|
21
|
Ashburner M, Ball CA, Blake JA, et al:
Gene Ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Q and Wang XJ: GOEAST: a web-based
software toolkit for Gene Ontology enrichment analysis. Nucleic
Acids Res. 36:W358–W363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kelder T, van Iersel MP, Hanspers K, et
al: WikiPathways: building research communities on biological
pathways. Nucleic Acids Res. 40:D1301–D1307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pico AR, Kelder T, van Iersel MP, Hanspers
K, Conklin BR and Evelo C: WikiPathways: pathway editing for the
people. PLoS Biol. 6:e1842008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duncan D, Prodduturi N and Zhang B:
WebGestalt2: an updated and expanded version of the Web-based Gene
Set Analysis Toolkit. BMC Bioinformatics. 11(Suppl 4): P102010.
View Article : Google Scholar
|
|
27
|
Jeanes A, Gottardi CJ and Yap AS:
Cadherins and cancer: how does cadherin dysfunction promote tumor
progression? Oncogene. 27:6920–6929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Do N, Zhao R, Ray K, et al: BMP4 is a
novel paracrine inhibitor of liver regeneration. Am J Physiol
Gastrointest Liver Physiol. 303:G1220–G1227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia Y, Babitt JL, Sidis Y, Chung RT and
Lin HY: Hemojuvelin regulates hepcidin expression via a selective
subset of BMP ligands and receptors independently of neogenin.
Blood. 111:5195–5204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giannelli G, Mazzocca A, Fransvea E, Lahn
M and Antonaci S: Inhibiting TGF-β signaling in hepatocellular
carcinoma. Biochim Biophys Acta. 1815:214–223. 2011.
|
|
31
|
Ikushima H and Miyazono K: TGFβ
signalling: a complex web in cancer progression. Nat Rev Cancer.
10:415–424. 2010.
|
|
32
|
Mackay F, Sierro F, Grey ST and Gordon TP:
The BAFF/APRIL system: an important player in systemic rheumatic
diseases. Curr Dir Autoimmun. 8:243–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jablonska E, Slodczyk B,
Wawrusiewicz-Kurylonek N, et al: Overexpression of B
cell-activating factor (BAFF) in neutrophils of oral cavity cancer
patients-preliminary study. Neoplasma. 58:211–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fragioudaki M, Tsirakis G, Pappa CA, et
al: Serum BAFF levels are related to angiogenesis and prognosis in
patients with multiple myeloma. Leuk Res. 36:1004–1008. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brown KA, Samarajeewa NU and Simpson ER:
Endocrine-related cancers and the role of AMPK. Mol Cell
Endocrinol. 366:170–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer - a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001.PubMed/NCBI
|
|
37
|
de Jong FA, de Jonge MJ, Verweij J and
Mathijssen RH: Role of pharmacogenetics in irinotecan therapy.
Cancer Lett. 234:90–106. 2006.PubMed/NCBI
|
|
38
|
Taves MD, Gomez-Sanchez CE and Soma KK:
Extra-adrenal glucocorticoids and mineralocorticoids: evidence for
local synthesis, regulation, and function. Am J Physiol Endocrinol
Metab. 301:E11–E24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rose AJ, Vegiopoulos A and Herzig S: Role
of glucocorticoids and the glucocorticoid receptor in metabolism:
insights from genetic manipulations. J Steroid Biochem Mol Biol.
122:10–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Amsterdam A, Tajima K and Sasson R:
Cell-specific regulation of apoptosis by glucocorticoids:
implication to their anti-inflammatory action. Biochem Pharmacol.
64:843–850. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Goldman DC, Bailey AS, Pfaffle DL, Al
Masri A, Christian JL and Fleming WH: BMP4 regulates the
hematopoietic stem cell niche. Blood. 114:4393–4401. 2009.
View Article : Google Scholar : PubMed/NCBI
|