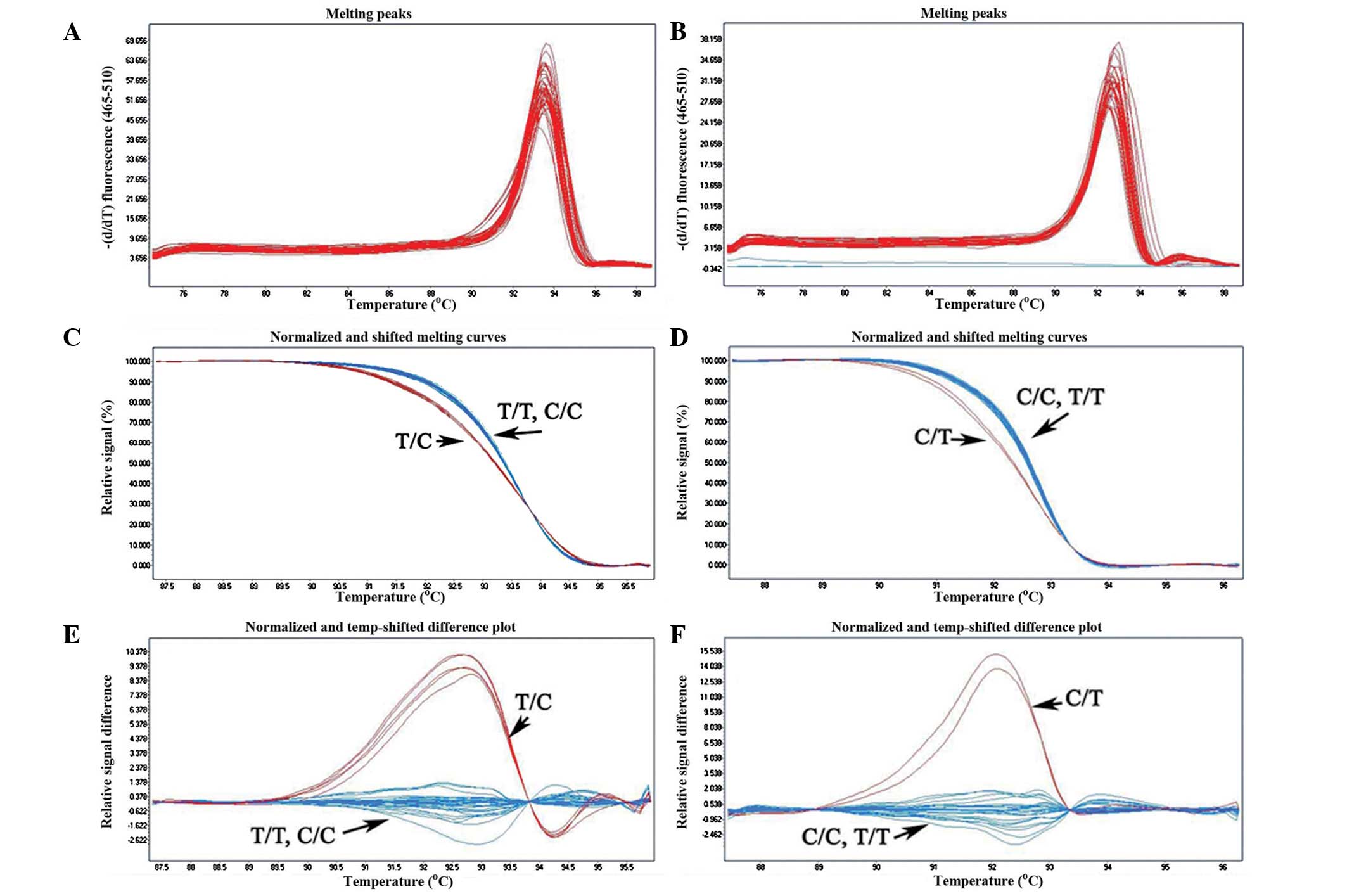
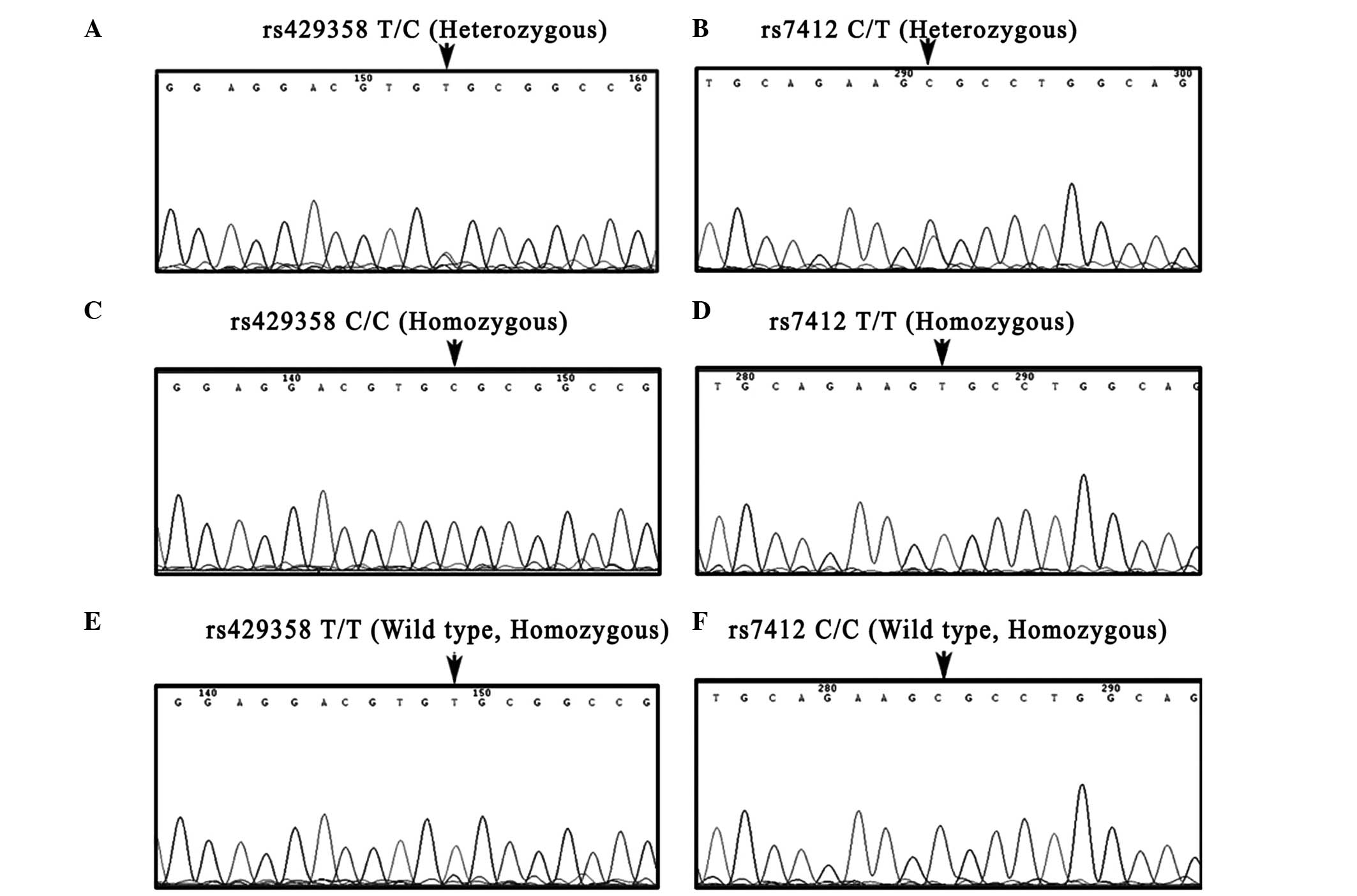
|
1
|
Kaneva AM, Bojko ER, Potolitsyna NN and
Odland JO: Plasma levels of apolipoprotein-E in residents of the
European North of Russia. Lipids Health Dis. 12:432013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahley RW, Innerarity TL, Rall SC Jr and
Weisgraber KH: Plasma lipoproteins: apolipoprotein structure and
function. J Lipid Res. 25:1277–1294. 1984.PubMed/NCBI
|
|
3
|
Eichner JE, Dunn ST, Perveen G, et al:
Apolipoprotein E polymorphism and cardiovascular disease: a HuGE
review. Am J Epidemiol. 155:487–495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pilia G, Chen WM, Scuteri A, et al:
Heritability of cardiovascular and personality traits in 6,148
Sardinians. PLoS Genet. 2:e1322006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin YW, Sun QQ, Zhang BB, et al:
Association between apolipoprotein E gene polymorphism and the risk
of coronary artery disease in Chinese population: evidence from a
meta-analysis of 40 studies. PLoS One. 8:e669242013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vaisi-Raygani A, Rahimi Z, Nomani H,
Tavilani H and Pourmotabbed T: The presence of apolipoprotein
epsilon4 and epsilon2 alleles augments the risk of coronary artery
disease in type 2 diabetic patients. Clin Biochem. 40:1150–1156.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lahiri DK, Sambamurti K and Bennett DA:
Apolipoprotein gene and its interaction with the environmentally
driven risk factors: molecular, genetic and epidemiological studies
of Alzheimer’s disease. Neurobiol Aging. 25:651–660. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerdes LU: The common polymorphism of
apolipoprotein E: geographical aspects and new pathophysiological
relations. Clin Chem Lab Med. 41:628–631. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Corbo RM and Scacchi R: Apolipoprotein E
(APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’
allele? Ann Hum Genet. 63:301–310. 1999. View Article : Google Scholar
|
|
10
|
El-Tagui MH, Hamdy MM, Shaheen IA, Agha H
and Abd-Elfatah HA: Apolipoprotein E gene polymorphism and the risk
of left ventricular dysfunction among Egyptian β-thalassemia major.
Gene. 524:292–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saidi S, Slamia LB, Ammou SB, Mahjoub T
and Almawi WY: Association of apolipoprotein E gene polymorphism
with ischemic stroke involving large-vessel disease and its
relation to serum lipid levels. J Stroke Cerebrovasc Dis.
16:160–166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamruecha W, Chansirikarnjana S, Nimkulrat
E, et al: Rapid detection of apolipoprotein E genotypes in
Alzheimer’s disease using polymerase chain reaction-single strand
conformation polymorphism. Southeast Asian J Trop Med Public
Health. 37:793–797. 2006.PubMed/NCBI
|
|
13
|
Calabretta A, Tedeschi T, Di Cola G, et
al: Arginine-based PNA microarrays for APOE genotyping. Mol
Biosyst. 5:1323–1330. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johansson Å, Enroth S, Palmblad M, et al:
Identification of genetic variants influencing the human plasma
proteome. Proc Natl Acad Sci USA. 110:4673–4678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Darawi MN, Ai-Vyrn C, Ramasamy K, et al:
Allele-specific polymerase chain reaction for the detection of
Alzheimer’s disease-related single nucleotide polymorphisms. BMC
Med Genet. 14:272013. View Article : Google Scholar
|
|
16
|
Pan M, Lin M, Yang L, et al:
Glucose-6-phosphate dehydrogenase (G6PD) gene mutations detection
by improved high-resolution DNA melting assay. Mol Biol Rep.
40:3073–3082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Furtado LV, Weigelin HC, Elenitoba-Johnson
KS and Betz BL: A multiplexed fragment analysis-based assay for
detection of JAK2 exon 12 mutations. J Mol Diagn. 15:592–599. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang YQ, Wu CJ, Yao M, Zhang YA and Zheng
LL: Study on the apo E gene polymorphism in Li population patients
with cardiovascular and cerebrovascular disease. Jianyan Yixue.
27:308–310. 3152012.(In Chinese).
|
|
19
|
Hu P, Qin YH, Lei FY, et al: Variable
frequencies of apolipoprotein E genotypes and its effect on serum
lipids in the Guangxi Zhuang and Han children. Int J Mol Sci.
12:5604–5615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kao JT, Tsai KS, Chang CJ and Huang PC:
The effects of apolipoprotein E polymorphism on the distribution of
lipids and lipoproteins in the Chinese population. Atherosclerosis.
114:55–59. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang KQ, Xie YH and He JL: The
investigation of apolipoprotein E polymorphism and genotype
distribution in the Chinese populations (Beijing and Tianjing).
Shengwu Huaxue Zazhi. 18:48–50. 1988.(In Chinese).
|
|
22
|
Mayila W, Fang MW, Cheng ZH and Qiu CC:
Polymorphism of apolipoprotein E gene and natural longevity in the
Xinjiang Uighur people: an association study. Zhonghua Yi Xue Yi
Chuan Xue Za Zhi. 22:462–463. 2005.(In Chinese). PubMed/NCBI
|
|
23
|
Huang H, Su M, Li X, et al: Y-chromosome
evidence for common ancestry of three Chinese populations with a
high risk of esophageal cancer. PLoS One. 5:e111182010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mas J, Yumbe A, Solé N, Capote R and
Cremades T: Prevalence, geographical distribution and clinical
manifestations of onchocerciasis on the Island of Bioko (Equatorial
Guinea). Trop Med Parasitol. 46:13–18. 1995.PubMed/NCBI
|
|
25
|
Calzada P, Suárez I, García S, et al: The
Fang population of Equatorial Guinea characterised by 15 STR-PCR
polymorphisms. Int J Legal Med. 119:107–110. 2005. View Article : Google Scholar
|
|
26
|
Wozniak MA, Faragher EB, Todd JA, et al:
Does apolipoprotein E polymorphism influence susceptibility to
malaria? J Med Genet. 40:348–351. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gerdes LU: The common polymorphism of
apolipoprotein E: geographical aspects and new pathophysiological
relations. Clin Chem Lab Med. 41:628–631. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kleinschmidt I, Sharp B, Benavente LE, et
al: Reduction in infection with Plasmodium falciparum one year
after the introduction of malaria control interventions on Bioko
Island, Equatorial Guinea. Am J Trop Med Hyg. 74:972–978.
2006.PubMed/NCBI
|
|
29
|
Vignali M, McKinlay A, LaCount DJ, et al:
Interaction of an atypical Plasmodium falciparum ETRAMP with human
apolipoproteins. Malar J. 7:2112008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wozniak MA, Riley EM and Itzhaki RF:
Apolipoprotein E polymorphisms and risk of malaria. J Med Genet.
41:145–146. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rougeron V, Woods CM, Tiedje KE, et al:
Epistatic interactions between apolipoprotein E and hemoglobin S
genes in regulation of malaria parasitemia. PLoS One. 8:e769242013.
View Article : Google Scholar : PubMed/NCBI
|