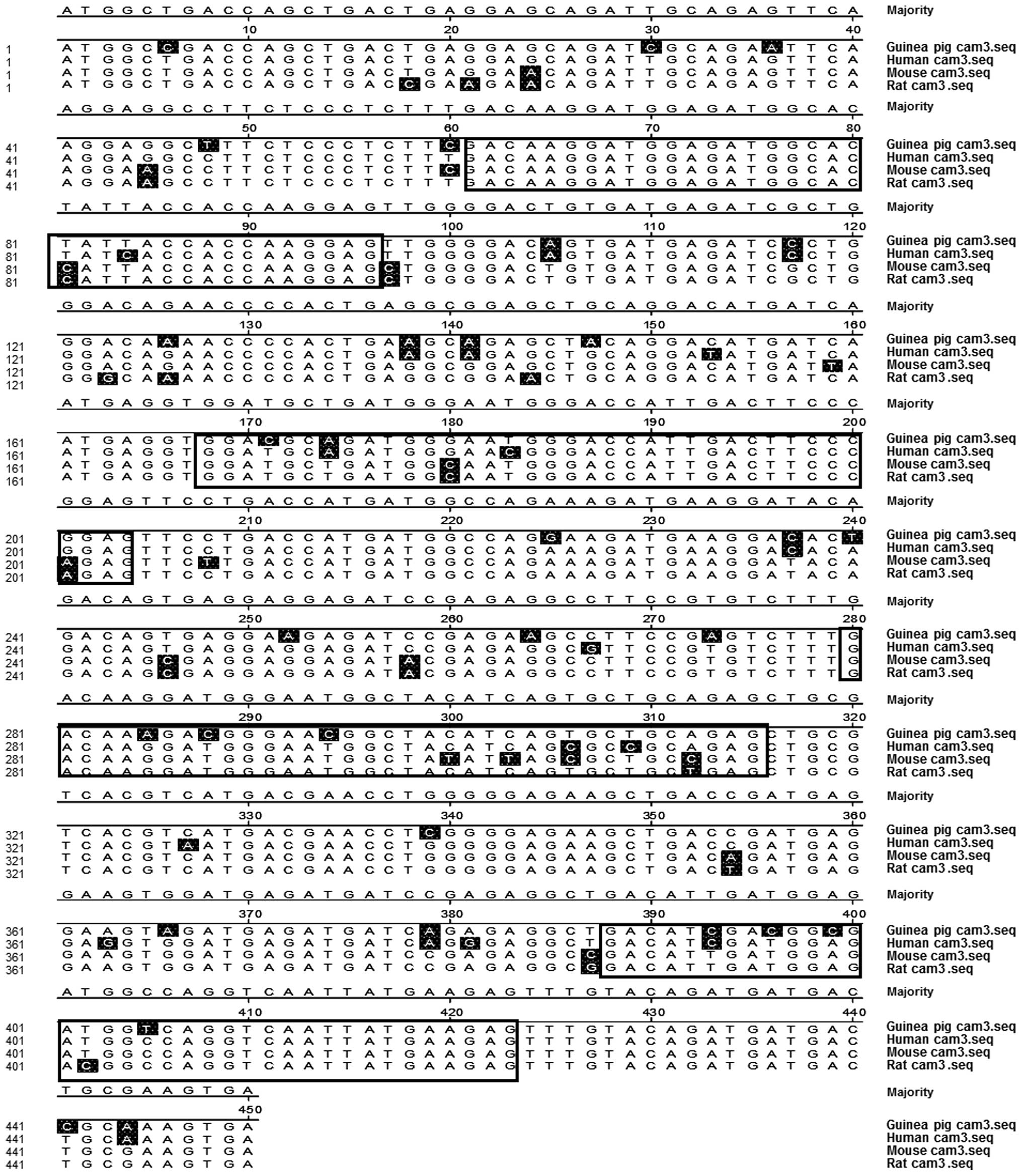
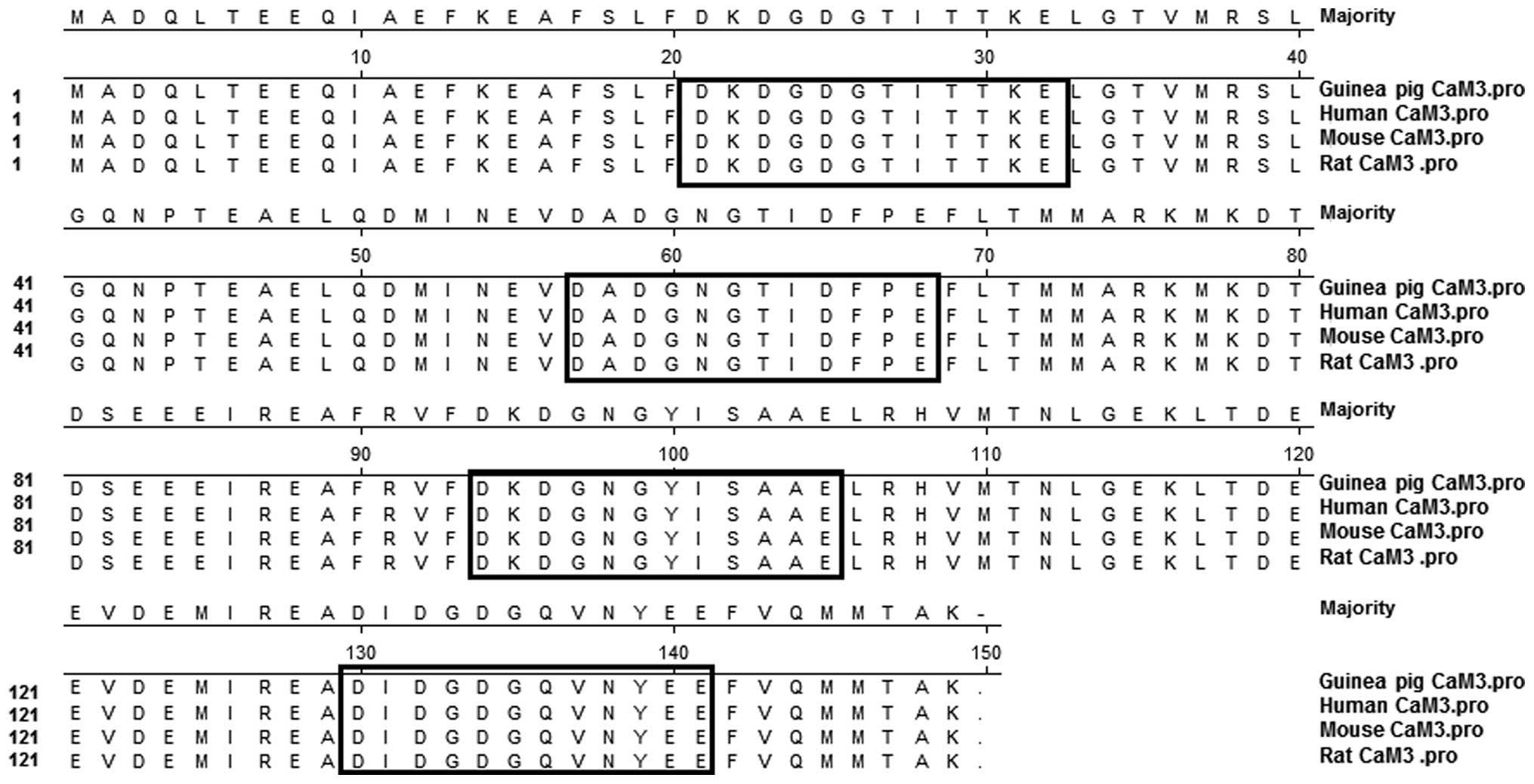
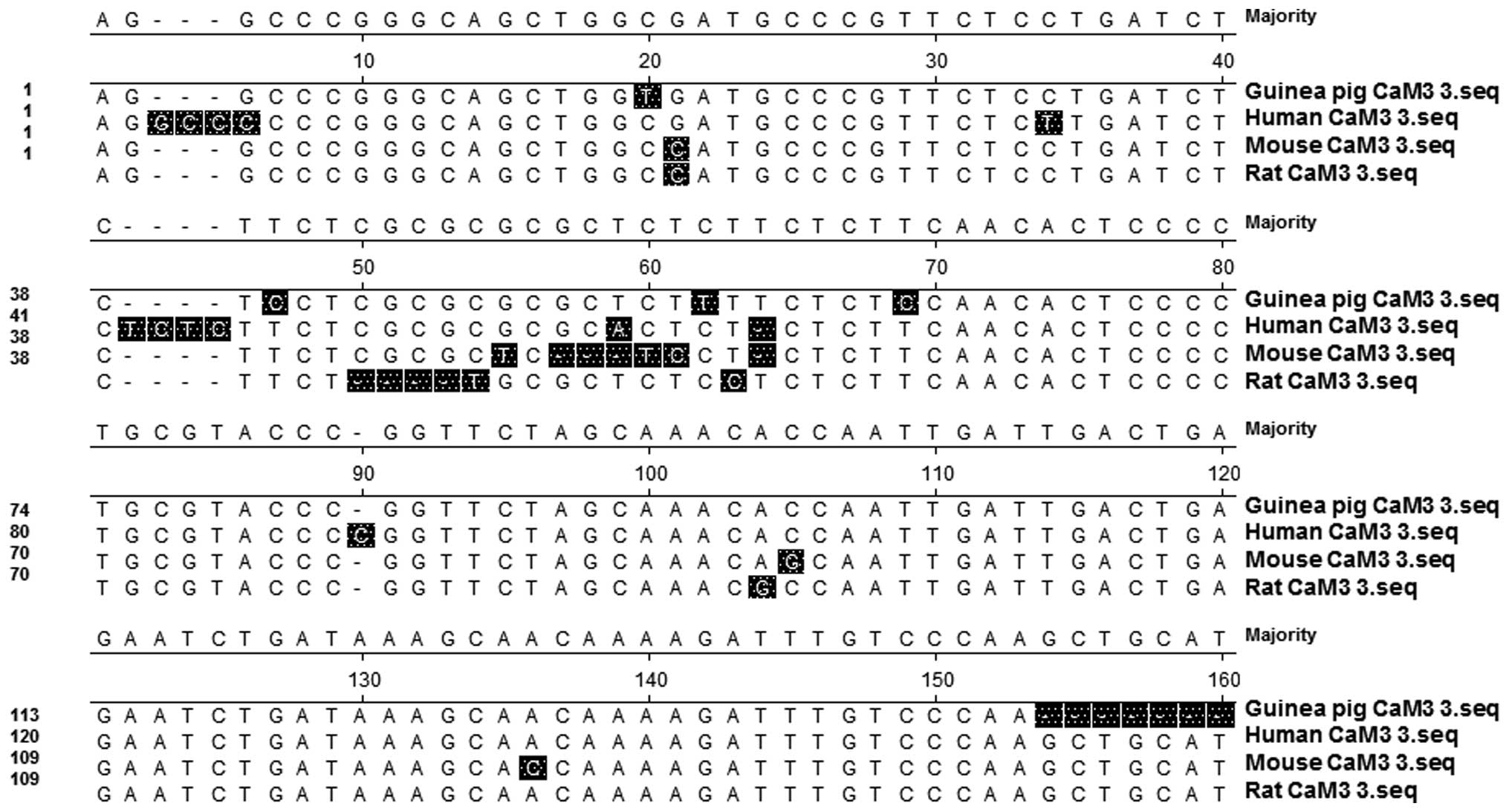
|
1
|
Ben-Johny M and Yue DT: Calmodulin
regulation (calmodulation) of voltage-gated calcium channels. J Gen
Physiol. 143:679–692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rasmussen CD and Means AR: Calmodulin is
involved in regulation of cell proliferation. EMBO J. 6:3961–3968.
1987.PubMed/NCBI
|
|
3
|
Deb TB, Coticchia CM and Dickson RB:
Calmodulin-mediated activation of Akt regulates survival of
c-Myc-overexpressing mouse mammary carcinoma cells. J Biol Chem.
279:38903–38911. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Day DH: CaMBOT: Profiling and
characterizing calmodulin-binding proteins. Cell Signal.
15:347–354. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yap KL, Kim J, Truong K, Sherman M, Yuan T
and Ikura M: Calmodulin target database. J Struct Funct Genomics.
1:8–14. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim J, Ghosh S, Nunziato DA and Pitt GS:
Identification of the components controlling inactivation of
voltage-gated Ca2+ channels. Neuron. 41:745–754. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zühlke RD, Pitt GS, Deisseroth K, Tsien RW
and Reuter H: Calmodulin supports both inactivation and
facilitation of L-type calcium channels. Nature. 399:159–162. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berrocal M, Sepulveda MR,
Vazquez-Hernandez M and Mata AM: Calmodulin antagonizes amyloid-β
peptides-mediated inhibition of brain plasma membrane
Ca2+-ATPase. Biochim Biophys Acta. 1822:961–969. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crotti L, Johnson CN, Graf E, et al:
Calmodulin mutations associated with recurrent cardiac arrest in
infants. Circulation. 127:1009–1017. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Babu YS, Bugg CE and Cook WJ: Structure of
calmodulin refined at 2.2 A resolution. J Mol Biol. 204:191–204.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein JP, Munjaal RP, Lagace L, Lai EC,
O'Malley BW and Means AR: Tissue-specific expression of a chicken
calmodulin pseudogene lacking intervening sequences. Proc Natl Acad
Sci USA. 80:6485–6489. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salas MA, Valverde CA, Sánchez G, et al:
The signalling pathway of CaMKII-mediated apoptosis and necrosis in
the ischemia/reperfusion injury. J Mol Cell Cardiol. 48:1298–1306.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Consolini AE and Bonazzola P: Energetics
of Ca2+ homeostasis during ischemia-reperfusion on
neonatal rat hearts under high-[K+] cardioplegia. Can J
Physiol Pharmacol. 86:866–879. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farah C, Meyer G, André L, et al: Moderate
exercise prevents impaired Ca2+ handling in heart of
CO-exposed rat: implication for sensitivity to
ischemia-reperfusion. Am J Physiol Heart Circ Physiol.
299:H2076–H2081. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wawrzynczak EJ and Perham RN: Isolation
and nucleotide sequence of a cDNA encoding human calmodulin.
Biochem Int. 9:177–185. 1984.PubMed/NCBI
|
|
16
|
SenGupta B, Friedberg F and
Detera-Wadleigh SD: Molecular analysis of human and rat calmodulin
complementary DNA clones. Evidence for additional active genes in
these species. J Biol Chem. 262:16663–16670. 1987.PubMed/NCBI
|
|
17
|
Fischer R, Koller M, Flura M, et al:
Multiple divergent mRNAs code for a single human calmodulin. J Biol
Chem. 263:17055–17062. 1988.PubMed/NCBI
|
|
18
|
Goldhagen H and Clarke M: Identification
of the single gene for calmodulin in Dictyostelium discoideum. Mol
Cell Biol. 6:1851–1854. 1986.PubMed/NCBI
|
|
19
|
Zimmer WE, Schloss JA, Silflow CD,
Youngblom J and Watterson DM: Structural organization, DNA sequence
and expression of the calmodulin gene. J Biol Chem.
263:19370–19383. 1988.PubMed/NCBI
|
|
20
|
Davis TN, Urdea MS, Masiarz FR and Thorner
J: Isolation of the yeast calmodulin gene: calmodulin is an
essential protein. Cell. 47:423–431. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takeda T and Yamamoto M: Analysis and in
vivo disruption of the gene coding for calmodulin in
Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 84:3580–3584.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei-xu H, Qin X, Zhu W, et al: Therapeutic
potential of anti-IL-1β IgY in guinea pigs with allergic asthma
induced by ovalbumin. Mol Immunol. 58:139–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Costa M, Dodds KN, Wiklendt L, Spencer NJ,
Brookes SJ and Dinning PG: Neurogenic and myogenic motor activity
in the colon of the guinea pig, mouse, rabbit, and rat. Am J
Physiol Gastrointest Liver Physiol. 305:G749–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Padilla-Carlin DJ, McMurray DN and Hickey
AJ: The guinea pig as a model of infectious diseases. Comp Med.
58:324–340. 2008.PubMed/NCBI
|
|
25
|
Shao D, Zhao M, Xu J, et al: The
individual N- and C-lobes of calmodulin tether to the Cav1.2
channel and rescue the channel activity from run-down in
ventricular myocytes of guinea-pig heart. FEBS Lett. 588:3855–3861.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng R, Xu J, Minobe E, et al: Adenosine
triphosphate regulates the activity of guinea pig Cav1.2 channel by
direct binding to the channel in a dose-dependent manner. Am J
Physiol Cell Physiol. 306:C856–863. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saitoh D, Asakura Y, Nkembo MK, et al:
Cloning and expression of calmodulin gene in Scoparia dulcis. Biol
Pharm Bull. 30:1161–1163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Al-Quraan NA, Locy RD and Singh NK:
Expression of calmodulin genes in wild type and calmodulin mutants
of Arabidopsis thaliana under heat stress. Plant Physiol Biochem.
48:697–702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Palfi A, Kortvely E, Fekete E, Kovacs B,
Varszegi S and Gulya K: Differential calmodulin gene expression in
the rodent brain. Life Sci. 70:2829–2855. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Toutenhoofd SL, Foletti D, Wicki R, Rhyner
JA, Garcia F, Tolon R and Strehler EE: Characterization of the
human CALM2 calmodulin gene and comparison of the transcriptional
activity of CALM1, CALM2 and CALM3. Cell Calcium. 23:323–338. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Toutenhoofd SL and Strehler EE: The
calmodulin multigene family as a unique case of genetic redundancy:
multiple levels of regulation to provide spatial and temporal
control of calmodulin pools? Cell Calcium. 28:83–96. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clapham DE: Calcium signaling. Cell.
131:1047–1058. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nojima H: Structural organization of
multiple rat calmodulin genes. J Mol Biol. 208:269–282. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berchtold MW, Egli R, Rhyner JA, Hameister
H and Strehler EE: Localization of the human bona fide calmodulin
genes CALM1, CALM2 and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3
and 19q13.2-q13.3. Genomics. 16:461–465. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Orojan I, Bakota L and Gulya K:
Differential calmodulin gene expression in the nuclei of the rat
midbrain-brain stem region. Acta Histochem. 108:455–462. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou LW, Moyer JA, Muth EA, Clark B,
Palkovits M and Weiss B: Regional distribution of calmodulin
activity in rat brain. J Neurochem. 44:1657–1662. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solà C, Tusell JM and Serratosa J:
Comparative study of the pattern of expression of calmodulin
messenger RNAs in the mouse brain. Neuroscience. 75:245–256. 1996.
View Article : Google Scholar : PubMed/NCBI
|