Introduction
Hepatocellular carcinoma (HCC), as the most common
primary cancer of the liver, contributes significantly to rates of
cancer-related morbidity and mortality worldwide. The annual
estimated incidence of new HCC cases is >0.5 million (1), and the highest incidence rates of HCC
(>20/100,000) have been reported in East and Southeast Asian
countries. In total, >80% of HCC cases occur in developing
countries (2). Thus, the diagnosis,
treatment and prevention of HCC represent significant challenges
worldwide. It is well established that the development of HCC is a
complex biological process, and the precise mechanism underlying
the occurrence of HCC is still not completely understood. Multiple
risk factors contribute to HCC susceptibility, such as chronic
hepatitis B virus (HBV) infection, aflatoxin B1 (AFB1) intake,
smoking and an excessive consumption of alcohol (3). In addition to these exogenous factors,
genetic polymorphisms play an important role in HCC
susceptibility.
Within recent decades, several studies have
evaluated and highlighted the association between DNA repair genes
and hepatocellular neoplasm susceptibility. Nucleotide excision
repair (NER) has been proposed to be the most versatile DNA repair
mechanism. Xeroderma pigmentosum complementary group D (XPD), also
known as excision repair cross-complementing group 2 (ERCC2), is a
gene located at chromosome 19q13.3 and encodes a type of NER enzyme
(4). Several important single
nucleotide polymorphisms have been identified in the XPD locus,
with high variant frequencies in exon 10 (rs1799793, codon 312 G→A,
Asp→Asn) and exon 23 (rs1052559, codon 751 A→C, Lys→Gln) (5). These two polymorphisms have been
largely investigated in genetic epidemiological studies, and
individuals with these polymorphisms exhibit a higher risk of
developing different types of cancer (6–8).
To date, molecular epidemiological studies have
investigated the effect of these two polymorphisms on the risk of
HCC (9–18); however, the results obtained in these
studies have been inconsistent, particularly in an Asian
population. In addition, the majority of studies have solely
examined the effect of XPD polymorphisms, while only a few studies
have explored the interaction of XPD with HBV and other
environmental factors. Furthermore, the small sample sizes and
missing genotypic data have potentially contributed to
false-positive or -negative findings. A meta-analysis is a useful
method to overcome the disadvantages of individual studies, thereby
increasing the statistical power and precision of the effect
estimates (19). The aim of the
present study, therefore, was to perform a meta-analysis of all
eligible studies to evaluate the association between
Lys751Gln and Asp312Asn polymorphisms and
hepatocellular neoplasm susceptibility, and to further explore the
interaction between XPD polymorphisms and HBV.
Materials and methods
Search strategy
All case-control studies of XPD polymorphisms and
HCC risk published in either the English or Chinese language up to
June 1, 2014 were identified by performing systematic searches in
PubMed, Embase, Google Scholar, the Chinese National Knowledge
Infrastructure (CNKI) and the Chinese Biomedicine databases. The
search terms used included: (‘XPD’ OR ‘ERCC2’ OR ‘DNA repair gene’
OR ‘xeroderma pigmentosum group D’) AND (‘polymorphism’ OR
‘variation’ OR ‘variant’ OR ‘genotype’ OR ‘genetic’) AND
(‘hepatocellular carcinoma’ OR ‘HCC’ OR ‘liver cancer’ OR ‘liver
tumour’ OR ‘liver neoplasms’ OR ‘hepatic tumour’). The references
of each identified article were also manually searched to identify
additional relevant publications. If more than one article was
published using the same case series, then only the study with the
largest sample size was included.
Inclusion criteria
A study was accepted for inclusion in the
meta-analysis if it met the following criteria: i) It assessed the
correlation between HCC and the XPD Lys751Gln or
Asp312Asn polymorphism in an Asian population; ii) it
employed a case-control design; iii) it provided sufficient data
regarding the genotypes (Gln/Gln, Lys/Lys, Gln/Lys, Asn/Asn,
Asp/Asp and Asn/Asp) to estimate an odds ratio (OR) with a 95%
confidence interval (95% CI); and iv) it clearly described the
method used to perform the HCC diagnoses and the sources of the
cases and controls. When multiple studies containing duplicate or
overlapping data were published by the same authors, the most
recent study with the largest participant population was
selected.
Data extraction
Literature searches and the identification of
eligible articles based on the inclusion criteria were conducted in
duplicate by two investigators using a standard protocol and
data-collection form (20). The
original extracted data were confirmed by another two
investigators, and any disagreement was resolved by discussion
among the four investigators. The following data were independently
extracted: Surname of the first author, ethnicity or country,
source of controls, genotyping methods, frequency and genotypes of
cases and controls, and the Hardy-Weinberg equilibrium (HWE) of the
controls.
Statistical analysis
The allelic frequency was initially calculated, and
the observed genotype frequencies of the XPD Lys751Gln
and Asp312Asn polymorphisms in the control group were
assessed for HWE using the χ2 test in each study. The
strength of the association between the XPD Lys751Gln
and Asp312Asn polymorphisms and HCC risk was assessed by
calculating ORs with 95% CIs. The overall pooled analysis was
performed for homozygote comparisons (Gln/Gln versus Lys/Lys and
Asn/Asn versus Asp/Asp), heterozygote comparisons (Lys/Gln versus
Lys/Lys and Asn/Asp versus Asp/Asp), the dominant model (Gln/Gln +
Lys/Gln versus Lys/Lys and Asn/Asn + Asp/Asn versus Asp/Asp), the
recessive model (Gln/Gln versus Lys/Gln + Lys/Lys and Asn/Asn
versus Asp/Asn + Asp/Asp) and allelic contrasts (Gln-allele versus
Lys-allele and Asn-allele versus Asp-allele). The significance of
the pooled ORs was determined using the Z-test, and P<0.05 was
considered to indicate statistical significance. The heterogeneity
of the studies was assessed using Cochran's Q-test and the
I2 test (21). If
P>0.1 or <50% the study was considered to lack heterogeneity,
in which case the pooled OR was calculated for each study using the
fixed-effects model. Otherwise, the random-effects model was used
(22). To assess the stability of
the results, a sensitivity analysis, in which each study in the
meta-analysis was systematically deleted to establish the impact of
each individual dataset on the overall pooled OR, was conducted.
Furthermore, the potential publication bias was evaluated through
visual inspection of the symmetry of the Begg's funnel plots. The
statistical analysis of the publication bias was conducted using
Egger's test. The meta-analysis was performed using Stata software
(version 12.0; StataCorp LP, College Station, TX, USA), and all the
P-values were two-sided.
Results
Characteristics of the studies
A flow chart of the exclusion/inclusion of the
studies in the meta-analysis is presented in Fig. 1. A total of 39 potentially relevant
publications up to June 1, 2014 were systematically identified
using the PubMed, Embase, Google Scholar and CNKI databases. Of
these publications, 23 studies (59.0%) were excluded due to a
failure to satisfy the inclusion criteria or to provide sufficient
information to determine whether the criteria were satisfied. An
additional four publications were excluded for one or more of the
following reasons: i) They did not examine the XPD
Lys751Gln or Asp312Asn polymorphisms; ii)
they did not provide allele frequencies, which were required for OR
calculation; iii) they deviated from the HWE; iv) they included a
family-based control or lacked a control group. Review articles and
bibliographies were manually searched to identify relevant studies.
A total of 11 studies (9–18; Yu, unpublished data) were ultimately
included in this meta-analysis based on the search strategy and
inclusion criteria. Four of the 11 studies examined both the XPD
Lys751Gln and Asp312Asn polymorphisms. In
total, 11 studies (2,852 HCC cases and 2,936 controls) for the XPD
Lys751Gln polymorphism (9–18; Yu, unpublished data) and
four studies (1,753 HCC cases and 1,914 controls) for the XPD
Asp312Asn polymorphism (11,13,15,17) were
retrieved in the final meta-analysis. The characteristics of the
included studies are listed in Tables
I and II.
 | Table I.Baseline characteristics and
methodological quality of the included studies investigating
Lys751Gln polymorphism. |
Table I.
Baseline characteristics and
methodological quality of the included studies investigating
Lys751Gln polymorphism.
|
|
|
|
|
| No. of cases | No. of
controls |
|
|---|
|
|
|---|
| First author, year
(ref.) | Ethnicity
(country) | Source of
controls | Genotyping
method | Cases/controls,
n/n | Lys/Lys | Lys/Glyn | Gln/Gln | Lys/Lys | Lys/Glyn | Gln/Gln | P-HWE |
|---|
| Xu, 2004 (6) | Asian (China) | PB | PCR-RFLP | 71/136 | 57 | 13 | 1 | 125 | 10 | 1 | 0.1348843 |
| Chen, 2005
(7) | Asian (China) | HB | PCR-RFLP | 570/381 | 496 | 72 | 2 | 322 | 55 | 4 | 0.3457913 |
| Xie, 2007 (8) | Asian (China) | HB | PCR-RFLP | 429/480 | 393 | 36 | 0 | 404 | 76 | 0 | 0.0596231 |
| Su, 2008 (9) | Asian (China) | HB | PCR-RFLP | 100/111 | 76 | 23 | 1 | 98 | 11 | 2 | 0.0244717 |
| Zeng, 2009
(10) | Asian (China) | HB | TaqMan-PCR | 300/312 | 263 | 32 | 5 | 270 | 39 | 3 | 0.2438713 |
| Cui, 2010 (11) | Asian (China) | HB | PCR-RFLP | 94/111 | 69 | 24 | 1 | 97 | 14 | 0 | 0.4782435 |
| Long, 2009
(12) | Asian (China) | HB | TaqMan-PCR | 618/712 | 272 | 222 | 124 | 464 | 187 | 61 |
1.361×10−9 |
| Yu,
2012a | Asian (China) | PB | PCR-RFLP | 76/80 | 32 | 26 | 18 | 54 | 20 | 6 | 0.0503993 |
| Hu, 2012 (13) | Asian (China) | PB | PCR-RFLP | 124/129 | 98 | 26 | 0 | 119 | 10 | 0 | 0.6469693 |
| Guo, 2012 (14) | Asian (China) | HB | PCR-CTPP | 410/410 | 190 | 183 | 37 | 233 | 159 | 18 | 0.1576467 |
| Gulnaz, 2013
(15) | Asian
(Pakistan) | HB | PCR-RFLP | 60/74 | 25 | 29 | 6 | 30 | 29 | 15 | 0.1161690 |
 | Table II.Baseline characteristics and
methodological quality of the included studies investigating XPD
Asp312Asn polymorphism. |
Table II.
Baseline characteristics and
methodological quality of the included studies investigating XPD
Asp312Asn polymorphism.
|
|
|
|
|
| No. of cases | No. of
controls |
|
|---|
|
|
|---|
| First author, year
(ref.) | Ethnicity
(country) | Source of
controls | Genotyping
method | Cases/controls,
n/n | Asp/Asp | Asp/Asn | Asn/Asn | Asp/Asp | Asp/Asn | Asn/Asn | P-HWE |
|---|
| Xie, 2007 (8) | Asian (China) | HB | PCR-RFLP | 425/480 | 404 | 21 | 0 | 445 | 35 | 0 | 0.4071126 |
| Zeng, 2009
(10) | Asian (China) | HB | PCR-RFLP | 300/312 | 204 | 92 | 4 | 252 | 54 | 6 | 0.1330516 |
| Long, 2009
(12) | Asian (China) | HB | TaqMan-PCR | 618/712 | 364 | 190 | 64 | 453 | 200 | 59 |
3.851×10−7 |
| Guo, 2012 (14) | Asian (China) | HB | PCR-CTPP | 410/410 | 260 | 107 | 43 | 282 | 96 | 32 |
2.548×10−7 |
Heterogeneity and sensitivity
analyses
The statistical heterogeneity of the XPD
Lys751Gln and Asp312Asn allelic contrast,
homozygote and heterozygote comparisons and dominant and recessive
genetic models were analysed for all studies and are shown in
Table III. With regard to the
heterogeneity of the XPD Lys751Gln and
Asp312Asn polymorphisms, no heterogeneity was found
among the studies for Asn/Asn versus Asp/Asp and the recessive
genetic model comparison (I2=0%); therefore,
fixed-effect models were used to analyse the OR. A random-effect
model was used in the pooled analysis of the other studies.
Following the test for heterogeneity, a sensitivity analysis was
performed to evaluate the stability of the results. The effect of
each study on the pooled OR was confirmed by repeating the
meta-analysis, while systematically omitting each study, one at a
time. The results were not materially altered and confirmed the
non-significant association between the XPD Lys751Gln
and Asp312Asn polymorphisms and HCC risk.
 | Table III.Heterogeneity and sensitivity
analyses and the quantitative synthesis of this meta-analysis. |
Table III.
Heterogeneity and sensitivity
analyses and the quantitative synthesis of this meta-analysis.
| A, XPD
Lys751Gln genetic comparison |
|---|
|
|---|
| | | Heterogeneity | |
|---|
|
|---|
| Comparison | OR (95% CI) | Z (P-value) | P-value | I2 | Analysis model |
|---|
| Gln-allele vs.
Lys-allele | 1.453
(1.032–2.046) | 2.14 (0.033) | <0.001 | 0.879 | Random |
| Gln/Gln vs.
Lys/Lys | 1.831
(1.001–3.349) | 1.96 (0.050) | 0.007 | 0.621 | Random |
| Lys/Gln vs.
Lys/Lys | 1.486
(1.044–2.114) | 2.20 (0.028) | <0.001 | 0.824 | Random |
| Gln/Gln + Lys/Gln
vs. Lys/Lys (dominant) | 1.540
(1.054–2.249) | 2.23 (0.026) | <0.001 | 0.864 | Random |
| Gln/Gln vs. Lys/Gln
+ Lys/Lys (recessive) | 1.603
(0.924–2.779) | 1.68 (0.093) | 0.018 | 0.568 | Random |
|
| B, XPD Asp312Asn
genetic comparison |
|
| | | Heterogeneity | |
|
| Comparison | OR (95% CI) | Z (P-value) | P-value | I2 | Analysis model |
|
| Asn-allele vs.
Asp-allele | 1.226
(0.965–1.557) | 1.67 (0.095) | 0.040 | 0.639 | Random |
| Asn/Asn vs.
Asp/Asp | 1.352
(1.010–1.808) | 2.03 (0.042) | 0.716 | <0.001 | Fixed |
| Asn/Asp vs.
Asp/Asp | 1.229
(0.857–1.762) | 1.12 (0.262) | 0.006 | 0.757 | Random |
| Asn/Asn + Asp/Asn
vs. Asp/Asp (dominant) | 1.249
(0.910–1.715) | 1.37 (0.169) | 0.013 | 0.723 | Random |
| Asn/Asn vs. Asp/Asn
+ Asp/Asp (recessive) | 1.250
(0.940–1.663) | 1.54 (0.125) | 0.515 | <0.001 | Fixed |
Quantitative synthesis
The main results of this pooled analysis are listed
in Table III. The association
between XPD polymorphism and HCC risk is shown in the form of
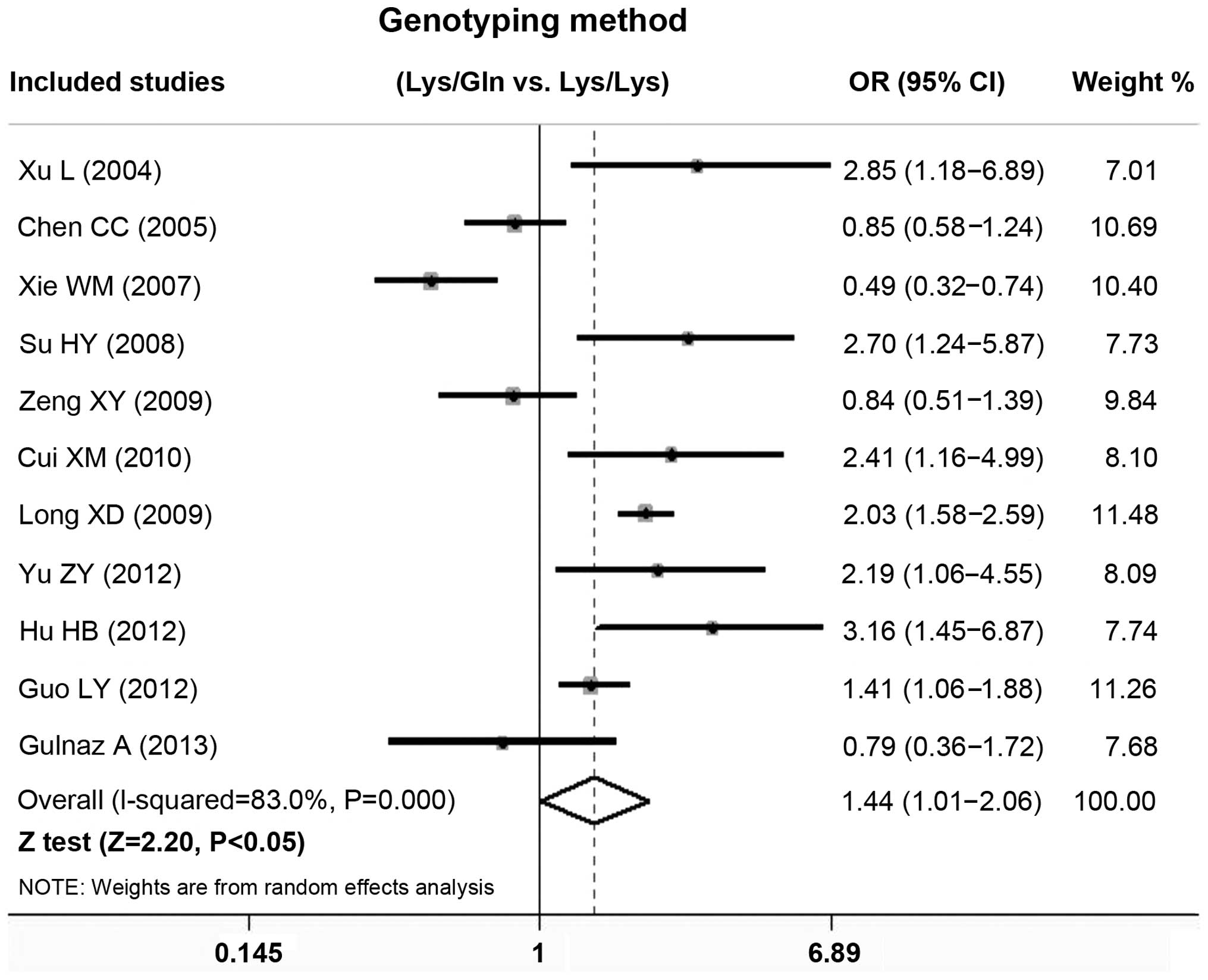
forest plots in Figs. 2 and 3. For the overall analysis, a significant
association between the risk of HCC and the variant genotypes of
the XPD Lys751Gln polymorphism was found in the
homozygote comparison (Gln/Gln versus Lys/Lys: OR, 1.831; 95% CI,
1.001–3.349), heterozygote comparison (Lys/Gln versus Lys/Lys: OR,
1.486; 95% CI, 1.044–2.114), dominant model (Gln/Gln + Lys/Gln
versus Lys/Lys: OR, 1.540; 95% CI, 1.054–2.249) and allelic
contrast (Gln-allele versus Lys-allele: OR, 1.453; 95% CI,
1.032–2.046). By contrast, no significant association was found in
the recessive model (Gln/Gln versus Lys/Gln + Lys/Lys: OR, 1.603;
95% CI, 0.924–2.779).
For the overall analysis, no significant association
between the risk of HCC and the variant genotypes of the XPD
Asp312Asn polymorphism was found in the heterozygote
comparison (Asn/Asp versus Asp/Asp: OR, 1.229; 95% CI,
0.857–1.762), dominant model (Asn/Asn + Asp/Asn versus Asp/Asp: OR,
1.249; 95% CI, 0.910–1.715), recessive model (Asn/Asn versus
Asp/Asn + Asp/Asp: OR, 1.250; 95% CI, 0.940–1.663) or allelic
contrast (Asn-allele versus Asp-allele: OR, 1.226; 95% CI,
0.965–1.557). By contrast, a significant association was found in
the homozygote comparison (Asn/Asn versus Asp/Asp: OR, 1.352; 95%
CI, 1.010–1.808).
Publication bias
Begg's funnel plots and Egger's test were used to
assess the publication bias for the reported comparisons of the XPD
Lys751Gln and Asp312Asn genotypes and HCC. As
shown in Fig. 4, no obvious
asymmetry in the comparison models was revealed for the XPD
Lys751Gln and Asp312Asn polymorphisms by
Begg's funnel plot. In addition, Egger's test was used to provide
statistical evidence of the funnel plot symmetry. These results
similarly did not show any evidence of publication bias (t=-0.38
and P=0.715 for Lys/Gln versus Lys/Lys; t=-0.13 and P=0.897 for the
dominant model of XPD Lys751Gln; t=-1.53 and P=0.170 for
the recessive model of XPD Lys751Gln; t=-0.59 and
P=0.569 for Gln-allele versus Lys-allele; t=-1.59 and P=0.358 for
Asn/Asn versus Asp/Asp; t=-0.27 and P=0.813 for Asn/Asp versus
Asp/Asp; t=-0.35 and P=0.761 for the dominant model of XPD
Asp312Asn; t=-0.89 and P=0.538 for the recessive model
of Lys751Gln; t=-0.44 and P=0.700 for Asn-allele versus
Asp-allele).
Gene-HBV interaction
In this meta-analysis, four of the studies (9,11,12,15)
analysed the gene-HBV interaction. Subgroup analyses were performed
based on whether or not the patients had chronic HBV infection, The
main results of this pooled analysis are shown in Table IV and Figs. 5 and 6, and the associations between XPD
Lys751Gln polymorphism/HBV status and HCC risk are
presented in the form of forest plots. No significant associations
were found in the HBV+ and HBV−
subgroups.
 | Table IV.Quantitative synthesis of subgroup
analyses concerning gene-HBV interaction for the XPD
Lys751Gln polymorphism. |
Table IV.
Quantitative synthesis of subgroup
analyses concerning gene-HBV interaction for the XPD
Lys751Gln polymorphism.
| Subgroup | Comparison | Studies (n) | OR (95% CI) |
|---|
| Total | Gln/Gln + Lys/Gln
vs. Lys/Lys (dominant) | 11 | 1.540
(1.054–2.249) |
|
HBV+ | Gln/Gln + Lys/Gln
vs. Lys/Lys (dominant) | 4 | 1.549
(0.527–4.554) |
|
HBV− | Gln/Gln + Lys/Gln
vs. Lys/Lys (dominant) | 4 | 1.058
(0.457–2.432) |
Discussion
Worldwide statistics indicate that liver cancer is
notably more common among men compared with women. In men, it is
the second leading cause of cancer death worldwide and in less
developed countries. An estimated 782,500 new liver cancer cases
and 745,500 deaths occurred worldwide during 2012, with China alone
accounting for ~50% of cases (23).
HCC is the most frequently occurring primary cancer
of the liver and the fifth most common solid tumour worldwide.
Between 500,000 and 1,000,000 new cases of HCC are believed to
occur each year, and HCC is estimated to cause 600,000 mortalities
globally each year (1). One
characteristic of HCC is the dysregulation of cell growth. HCC has
a complex, multistep and heterogeneous malignant tumourigenesis.
The pathogenesis of HCC involves a host of genetic and
environmental factors and the modulation of molecular signalling
pathways that have been implicated in the malignant transformation
of hepatocytes and tumour progression (24).
XPD is a DNA repair gene that is also known as ERCC2
(25). XPD plays a key role in NER,
a process that is crucial in the elimination of specific DNA
cross-links, ultraviolet photo-lesions and bulky chemical adducts
(26,27). XPD is a DNA-dependent ATPase/helicase
that is associated with the transcription factor IIH complex and
acts to perform a pivotal function in the NER pathway. XPD is
involved in the opening of the DNA duplex, which is necessary for
the excision of the fragment of DNA containing the damaged base
(28,29).
Numerous meta-analyses have been performed to
investigate the association between XPD polymorphism and the risk
of different types of cancer, e.g., prostate (30–32),
lung (33–35), gastric (8,36),
breast (7,37,38),
head and neck (39,40) and oesophageal (41,42). In
addition, several studies investigating XPD polymorphisms and HCC
susceptibility have been reported. Due to differences in district,
ethnicity and genotyping method, as well as an insufficient sample
size, however, the association between XPD polymorphism and HCC
risk remains conflicting and contradictory. Furthermore, few
meta-analyses have investigated the association between the XPD
Lys751Gln and Asp312Asn polymorphisms and HCC
risk, indicating the necessity for a meta-analysis to provide a
quantitative approach for combining different results. To derive a
more precise estimation of the association, we performed a
meta-analysis of 11 studies, including 2,852 cases and 2,936
controls.
To the best of our knowledge, this study has been
the first meta-analysis to investigate the association between the
Lys751Gln and Asp312Asn polymorphisms and HCC
risk among Asians. The meta-analysis included 2,852 cases and 2,936
controls from 11 case-control studies. A comparison of the pooled
ORs revealed a significantly increased risk in the homozygote
comparisons (Gln/Gln versus Lys/Lys and Asn/Asn versus Asp/Asp),
heterozygote comparison (Lys/Gln versus Lys/Lys), the dominant
model (Gln/Gln + Lys/Gln versus Lys/Lys) and allelic contrast
(Gln-allele versus Lys-allele). These results indicated that the
XPD Lys751Gln and Asp312Asn polymorphisms
could increase the risk of HCC.
It is well established that chronic HBV infection
plays a specific role in the development of HCC. The present
meta-analysis further explored this interaction between HBV and
HCC. The results showed that no significantly increased risks were
associated with the XPD Lys751Gln gene polymorphisms and
HCC risk in the HBV+ and HBV− subgroups;
however, only a small number of studies have examined the
association between the XPD Lys751Gln polymorphism and
HCC risk in HBV+ and HBV− patients.
Furthermore, the P-value of the Q-test for heterogeneity was
significant. Considering the limited number of studies and P-value
of the Q-test for heterogeneity in this meta-analysis, the present
results should be interpreted with caution.
A number of limitations to this meta-analysis should
be considered in the interpretation of the results. First, a common
limitation of meta-analyses is study heterogeneity. Heterogeneity
is often caused by variations in the environmental and genetic
backgrounds of the study participants, which is unavoidable when
assessing several studies. Evidence of study heterogeneity was
found in the present meta-analysis, presumably due to the fact that
the number of included studies was small, and the frequency of the
demographic variables for the cases and controls in one study was
not matched in age, gender or histological type, among other
factors. A second limitation was that the control groups were
selected from different populations. In certain studies, the
controls were healthy individuals, while in other studies the
reference group was selected from hospital patients without organic
gastric cancer; thus, there was a possibility of non-differential
misclassification bias, since these studies may have included
control groups with different risks of developing HCC. Thirdly, the
present results were based on single-factor estimates without
adjustment for other risk factors, including AFB1 intake, excessive
consumption of alcohol and other lifestyle factors. Finally, the
XPD gene could influence HCC susceptibility through interaction
with other genes; however, no gene-gene interaction analyses were
performed in this study.
In conclusion, the present meta-analysis provides
limited evidence to support the association of XPD
Lys751Gln and Asp312Asn polymorphisms with
HCC risk. No significantly increased risks were found to be
associated with the XPD Lys751Gln gene polymorphism and
HCC risk between the HBV+ and HBV− subgroups.
Large-scale studies with consideration for
gene-gene/gene-environment interactions should be performed to
further investigate the findings of the present study.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81260675) and Guangxi Natural
Science Foundation (no. 2013GXNSFAA019154) and the Project of
Outstanding Young Teachers Training in Higher Education
Institutions of Guangxi.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nordenstedt H, White DL and El-Serag HB:
The changing pattern of epidemiology in hepatocellular carcinoma.
Dig Liver Dis. 42 Suppl 3:S206–S214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao J, Xie L, Yang WS, et al: Risk factors
of hepatocellular carcinoma-current status and perspectives. Asian
Pac J Cancer Prev. 13:743–752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Flejter WL, McDaniel LD, Johns D,
Friedberg EC and Schultz RA: Correction of xeroderma pigmentosum
complementation group D mutant cell phenotypes by chromosome and
gene transfer: Involvement of the human ERCC2 DNA repair gene. Proc
Natl Acad Sci USA. 89:261–265. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen MR, Jones IM and Mohrenweiser H:
Nonconservative amino acid substitution variants exist at
polymorphic frequency in DNA repair genes in healthy humans. Cancer
Res. 58:604–608. 1998.PubMed/NCBI
|
|
6
|
Seker H, Butkiewicz D, Bowman ED, et al:
Functional significance of XPD polymorphic variants: Attenuated
apoptosis in human lymphoblastoid cells with the XPD 312 Asp/Asp
genotype. Cancer Res. 61:7430–7434. 2001.PubMed/NCBI
|
|
7
|
Pabalan N, Francisco-Pabalan O, Sung L,
Jarjanazi H and Ozcelik H: Meta-analysis of two ERCC2 (XPD)
polymorphisms, Asp312Asn and Lys751Gln, in breast cancer. Breast
Cancer Res Treat. 124:531–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin QH, Liu C, Hu JB, Meng RR, Li L and
Wang YJ: XPD Lys751Gln and Asp312Asn polymorphisms and gastric
cancer susceptibility: A meta-analysis of case-control studies.
Asian Pac J Cancer Prev. 14:231–236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu L, Wu YQ, Jing Y, et al: A case-control
study on polymorphism of DNA repair gene XPD and susceptibility to
hepatocellular carcinoma. Tumor. 24:5265292004.(In Chinese).
|
|
10
|
Chen CC, Yang SY, Liu CJ, et al:
Association of cytokine and DNA repair gene polymorphisms with
hepatitis B-related hepatocellular carcinoma. Int J Epidemiol.
34:1310–1318. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie WM: Association between the nucleotide
excision repair (NER) genes polymorphisms and genetic
susceptibility and clinical phenotype of hepatocellular carcinoma.
University of Guangxi Medical; 2007, (In Chinese).
|
|
12
|
Su HY: A case-control study on the
association between single nucleotide polymorphism of DNA injury
repair gene XPD, XRCC1 and hepatocellular carcinoma. University of
China Medical; 2008, (In Chinese).
|
|
13
|
Zeng XY, Qiu XQ, Ji L and Yu HP: Study on
the relationship between hepatocellular carcinoma and the
interaction between polymorphisms in DNA repair gene XPD and
environmental factors. Zhonghua Liu Xing Bing Xue Za Zhi.
30:7027052009.(In Chinese). PubMed/NCBI
|
|
14
|
Cui ZM and Su HY: A case-control study on
the polymorphism of gene XPD and the susceptibility of primary
hepatic carcinoma. Med J Chin People Health. 22:9129142010.(In
Chinese).
|
|
15
|
Long XD, Ma Y, Zhou YF, et al: XPD codon
312 and 751 polymorphisms, and AFB1 exposure, and hepatocellular
carcinoma risk. BMC Cancer. 9:4002009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu HB: A study on relationship between
familial clustering of hepatocellular carcinoma and such factors as
polymorphism of XPD genes, etc. in Guangxi popution. University of
Guangxi Medical; 2012, (In Chinese).
|
|
17
|
Guo LY, Jin XP, Niu W, Li XF, Liu BH and
Wang YL: Association of XPD and XRCC1 genetic polymorphisms with
hepatocellular carcinoma risk. Asian Pac J Cancer Prev.
13:4423–4426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gulnaz A, Sayyed AH, Amin F, et al:
Association of XRCC1, XRCC3, and XPD genetic polymorphism with an
increased risk of hepatocellular carcinoma because of the hepatitis
B and C virus. Eur J Gastroenterol Hepatol. 25:166–179. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Li Z, Feng L, Guo W and Zhang S:
Polymorphisms of DNA repair gene XRCC1 and hepatocellular carcinoma
risk among East Asians: A meta-analysis. Tumour Biol. 34:261–269.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang SX, Wu FX, Luo M, et al: The
glutathione S-Transferase P1 341C>T polymorphism and cancer
risk: A meta-analysis of 28 case-control studies. PLoS One.
8:e567222013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Torre LA, Bray F, Siegel RL, et al: Global
Cancer Statistics, 2012. CA Cancer J Clin. 61:86–108. 2015.
|
|
24
|
Llovet JM and Bruix J: Novel advancements
in the management of hepatocellular carcinoma in 2008. J Hepatol.
48 Suppl 1:S20–S37. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiao L, Hassan MM, Bondy ML, Abbruzzese
JL, Evans DB and Li D: The XPD Asp312Asn and Lys751Gln
polymorphisms, corresponding haplotype, and pancreatic cancer risk.
Cancer Lett. 245:61–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Au WW, Navasumrit P and Ruchirawat M: Use
of biomarkers to characterize functions of polymorphic DNA repair
genotypes. Int J Hyg Environ Health. 207:301–313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cleaver JE: Common pathways for
ultraviolet skin carcinogenesis in the repair and replication
defective groups of xeroderma pigmentosum. J Dermatol Sci. 23:1–11.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Benhamou S and Sarasin A: ERCC2/XPD gene
polymorphisms and cancer risk. Mutagenesis. 17:463–469. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Manuguerra M, Saletta F, Karagas MR, et
al: XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the
risk of cancer: A HuGE review. Am J Epidemiol. 164:297–302. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lavender NA, Komolafe OO, Benford M, et
al: No association between variant DNA repair genes and prostate
cancer risk among men of African descent. Prostate. 70:113–119.
2010.PubMed/NCBI
|
|
31
|
Zhu H, Cao S, Liu Y, Ding X, Wu Q and Ma
H: Genetic polymorphisms of xeroderma pigmentosum group D and
prostate cancer risk: A meta-analysis. J Cancer Res Ther.
9:187–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu S, Zhang H, Tang Y and Wang J:
Polymorphisms in XPD and hOGG1 and prostate cancer risk: A
meta-analysis. Urol Int. 89:233–240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feng Z, Ni Y, Dong W, Shen H and Du J:
Association of ERCC2/XPD polymorphisms and interaction with tobacco
smoking in lung cancer susceptibility: A systemic review and
meta-analysis. Mol Biol Rep. 39:57–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mei CR, Luo M, Li HM, Deng WJ and Zhou QH:
DNA Repair Gene Polymorphisms in the Nucleotide Excision Repair
Pathway and Lung Cancer Risk: A Meta-analysis. Chin J Cancer Res.
23:79–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhan P, Wang Q, Wei SZ, et al: ERCC2/XPD
Lys751Gln and Asp312Asn gene polymorphism and lung cancer risk: A
meta-analysis involving 22 case-control studies. J Thorac Oncol.
5:1337–1345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xue H, Lu Y, Lin B, Chen J, Tang F and
Huang G: The effect of XPD/ERCC2 polymorphisms on gastric cancer
risk among different ethnicities: A systematic review and
meta-analysis. PLoS One. 7:e434312012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang Z, Li C, Xu Y, Cai S and Wang X:
Associations between XPD polymorphisms and risk of breast cancer: A
meta-analysis. Breast Cancer Res Treat. 123:203–212. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiu LX, Yao L, Zhang J, et al: XPD
Lys751Gln polymorphism and breast cancer susceptibility: A
meta-analysis involving 28,709 subjects. Breast Cancer Res Treat.
124:229–235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu YY, Yuan H, Jiang GB, et al:
Associations between XPD Asp312Asn polymorphism and risk of head
and neck cancer: A meta-analysis based on 7,122 subjects. PLoS One.
7:e352202012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yuan H, Niu YM, Wang RX, Li HZ and Chen N:
Association between XPD Lys751Gln polymorphism and risk of head and
neck cancer: A meta-analysis. Genet Mol Res. 10:3356–3364. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Duan XL, Gong H, Zeng XT, et al:
Association between XPD Asp312Asn polymorphism and esophageal
cancer susceptibility: A meta-analysis. Asian Pac J Cancer Prev.
13:3299–3303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu XB, Dai LP, Wang YP, Wang KJ and Zhang
JY: DNA repair gene xeroderma pigmentosum group D 751 polymorphism
and the risk on esophageal cancer: A meta-analysis. Zhonghua Liu
Xing Bing Xue Za Zhi. 30:2812852009.(In Chinese). PubMed/NCBI
|