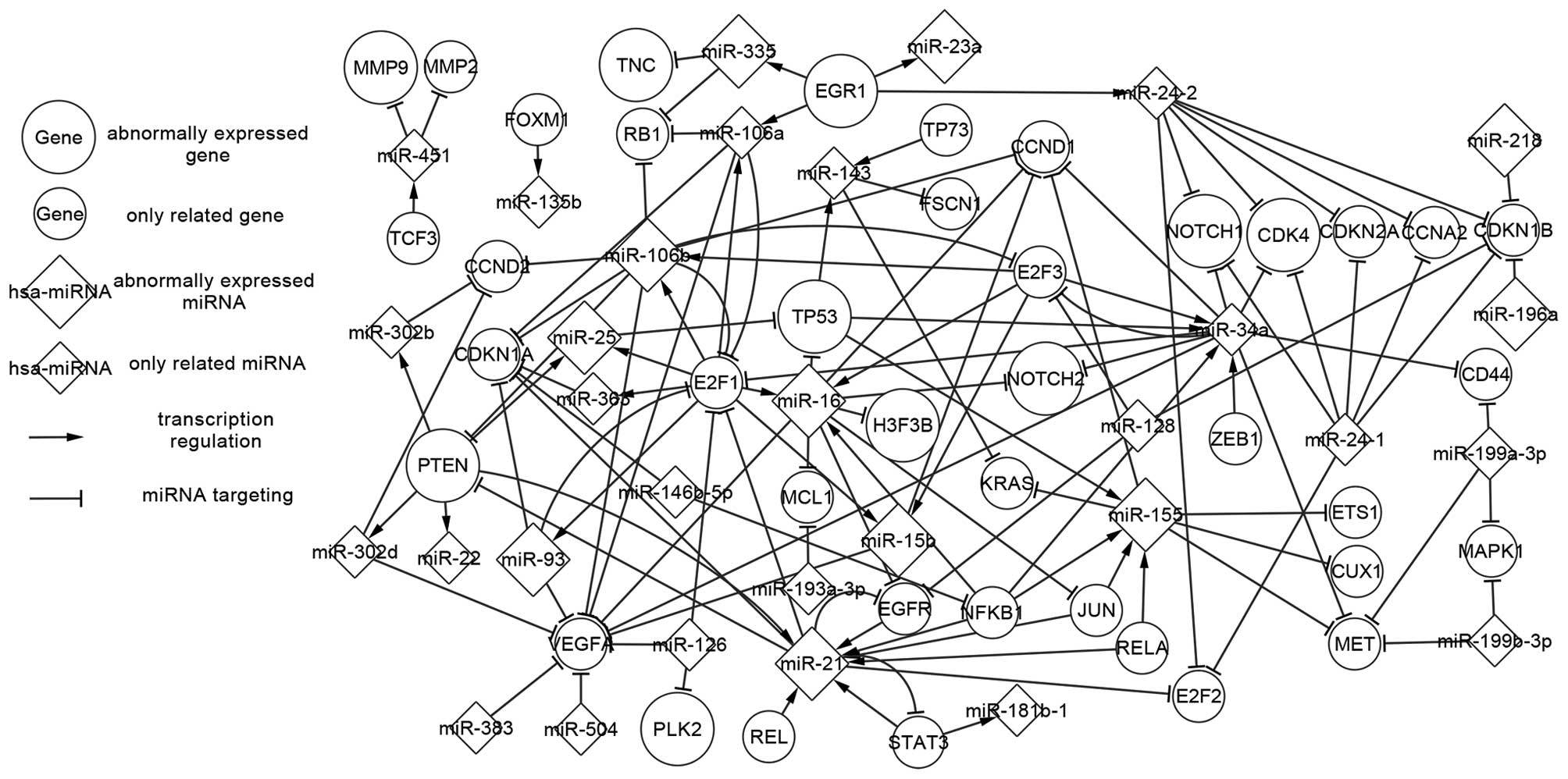
|
1
|
Landis SH, Murray T, Bolden S and Wingo
PA: Cancer statistics, 1999. CA Cancer J Clin. 49:8–31. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rao SA, Santosh V and Somasundaram K:
Genome-wide expression profiling identifies deregulated miRNAs in
malignant astrocytoma. Mod Pathol. 23:1404–1417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Polishsetty RV, Gautam P, Gupta MK, Sharma
R, Uppin MS, Challa S, Ankathi P, Purohit AK, Renu D, Harsha HC, et
al: Heterogeneous nuclear ribonucleoproteins and their interactors
are a major class of deregulated proteins in anaplastic
astrocytoma: A grade III malignant glioma. J Proteome Res.
12:3128–3138. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tanwar MK, Glibert MR and Holland EC: Gene
expression microarray analysis reveals YKL-40 to be a potential
serum marker for malignant character in human glioma. Cancer Res.
62:4364–4368. 2002.PubMed/NCBI
|
|
5
|
Ugur HC, Taspinar M, Ilgaz S, Sert F,
Canpinar H, Rey JA, Castresana JS and Sunguroglu A:
Chemotherapeutic resistance in anaplastic astrocytoma cell lines
treated with a temozolomide-lomeguatrib combination. Mol Biol Rep.
41:697–703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tran DH, Satou K, Ho TB and Pham TH:
Computational discovery of miR TF regulatory modules in human
genome. Bioinformation. 4:371–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:W1785–W1786. 2008. View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang
Q, Zhang Y, Wang R, Xue L, Wang S, et al: MiR-106a-5p inhibits the
proliferation and migration of astrocytoma cells and promotes
apoptosis by targeting FASTK. PLoS One. 8:e723902013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, Li J, Li S, Chen L, Ding H, et al: Intronic miR-301 feedback
regulates its host gene, ska2, in A549 cells by targeting MEOX2 to
affect ERK/CREB pathways. Biochem Biophys Res Commun. 396:978–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sandro PC, Jose DA, Carlos AC, Cristovam
SN, Stela VP, Sonia MM, Marcelo JDS and Euclides TDR: Evaluation of
the microvascular density in astrocytomas in adults correlated
using SPECT-MIBI. Exp Ther Med. 1:293–299. 2010.
|
|
14
|
Wang K, Xu Z, Wang N, Xu T and Zhu M:
MicroRNA and gene networks in human diffuse large B-cell lymphoma.
Oncol Lett. 8:2225–2232. 2014.PubMed/NCBI
|
|
15
|
Zhu MH, Xu ZW, Wang KH, Wang N, Zhu MY and
Wang S: MicroRNA and gene networks in human Hodgkin's lymphoma. Mol
Med Rep. 8:1747–1754. 2013.PubMed/NCBI
|
|
16
|
Killela PJ, Pirozzi CJ, Reitman ZJ, Jones
S, Rasheed BA, Lipp E, Friedman H, Friedman AH, He Y, McLendon RE,
et al: The genetic landscape of anaplastic astrocytoma. Oncotarget.
5:1452–1457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guan YL, Mizoguchi M, Yoshimoto K, Hata N,
Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al:
MiRNA-196 is upregulated in glioblastoma but not in anaplastic
astrocytoma and has prognostic significance. Clin Cancer Res.
16:4289–4297. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Xu ZW, Wang KH, Wang N, Li DQ and
Wang S: Networks of microRNAs and genes in retinoblastomas. Asian
Pac J Cancer Prev. 14:6631–6636. 2013. View Article : Google Scholar
|
|
19
|
Chekmenev DS, Haid C and Kel AE: P-Match:
Transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Papadopoulos GL, Reczko M, Simossis VA,
Sethupathy P and Hatzigeorgiou AG: The database of experimentally
supported targets: A functional update of TarBase. Nucleic Acids
Res. 37:D155–D158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Lu M, Qiu CX and Cui QH: TransmiR:
A transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Safran M, Dalah I, Alexander J, Rosen N,
Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al:
GeneCards Version 3: The human gene integrator. Database (Oxford).
2010:baq0202010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC genome browser database: Update 2011. Nucleic Acids
Res. 39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smith JS, Tachibana I, Passe SM, Huntley
BK, Borell TJ, Iturria N, O'Fallon JR, Schaefer PL, Scheithauer BW,
James CD, et al: PTEN mutation, EGFR amplification and outcome in
patients with anaplastic astrocytoma and glioblastoma multiforme. J
Natl Cancer Inst. 93:1246–1256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang CL: Genome-wide oligonucleotide
microarray analysis of gene-expression profiles of Taiwanese
patients with anaplastic astrocytoma and glioblastoma multiforme. J
Biomol Screen. 13:912–921. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, et al: Targeting oncogenic miR-335
inhibits growth and invasion of malignant astrocytoma cells. Mol
Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anselmo NP, Rey JA, Almeida LO, Custodio
AC, Almeida JR, Clara CA, Santos MJ and Casartelli C: Concurrent
sequence variation of TP53 and TP73 genes in anaplastic
astrocytoma. Genet Mol Res. 8:1257–1263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ushio Y, Tada K, Shiraishi S, Kamiryo T,
Shinojima N, Kochi M and Saya H: Correlation of molecular genetic
analysis of p53, MDM2, P16, PTEN and EGFR and survival of patients
with anaplastic astrocytoma and glioblastoma. Front Biosci.
8:e281–e288. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhi F, Chen X, Wang SN, Xia X, Shi Y, Guan
W, Shao N, Qu H, Yang C, Zhang Y, et al: The use of hsa-miR-21,
hsa-miR-181b and hsa-miR-106a as prognostic indicators of
astrocytoma. Eur J Cancer. 46:1640–1649. 2010. View Article : Google Scholar : PubMed/NCBI
|