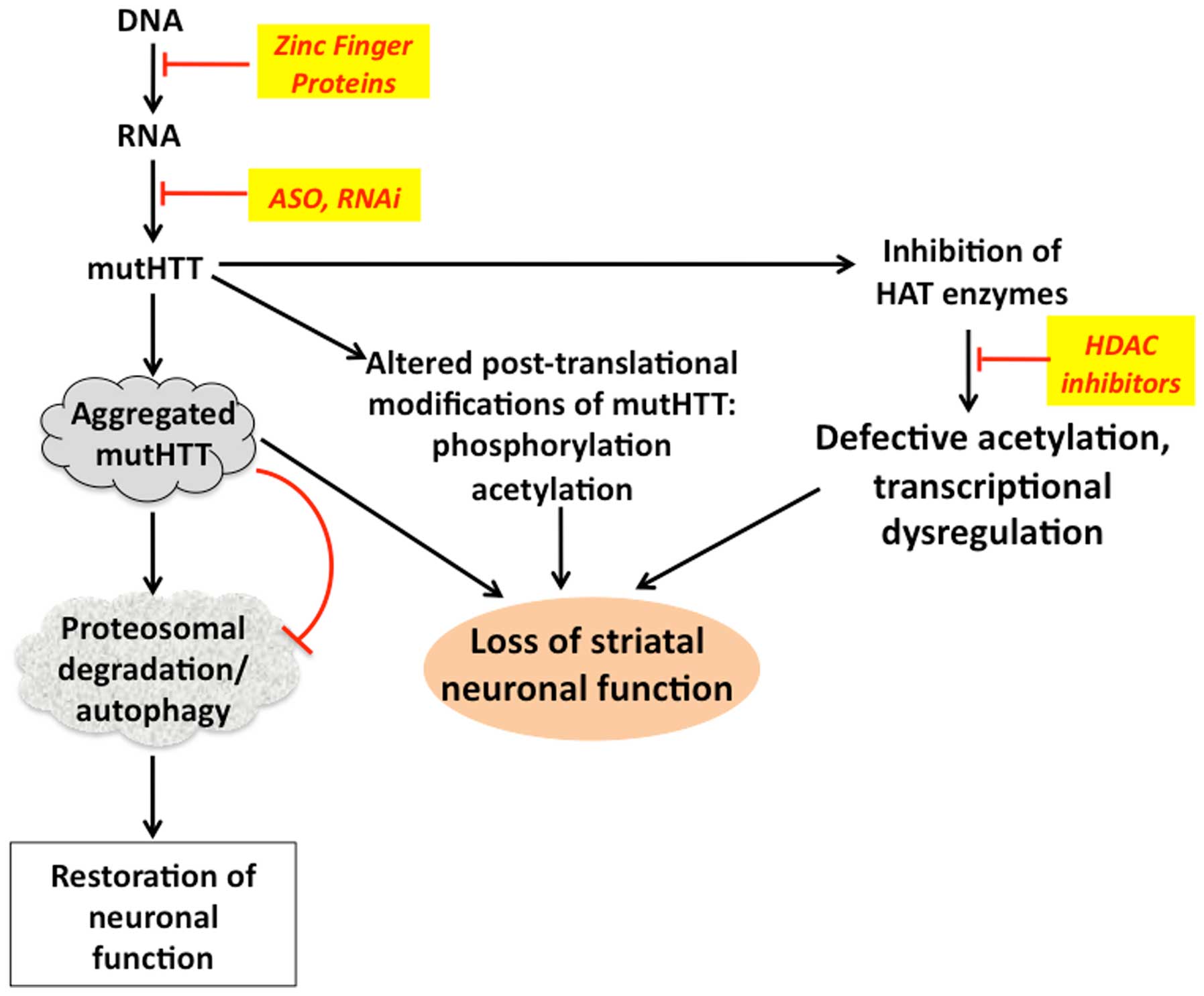
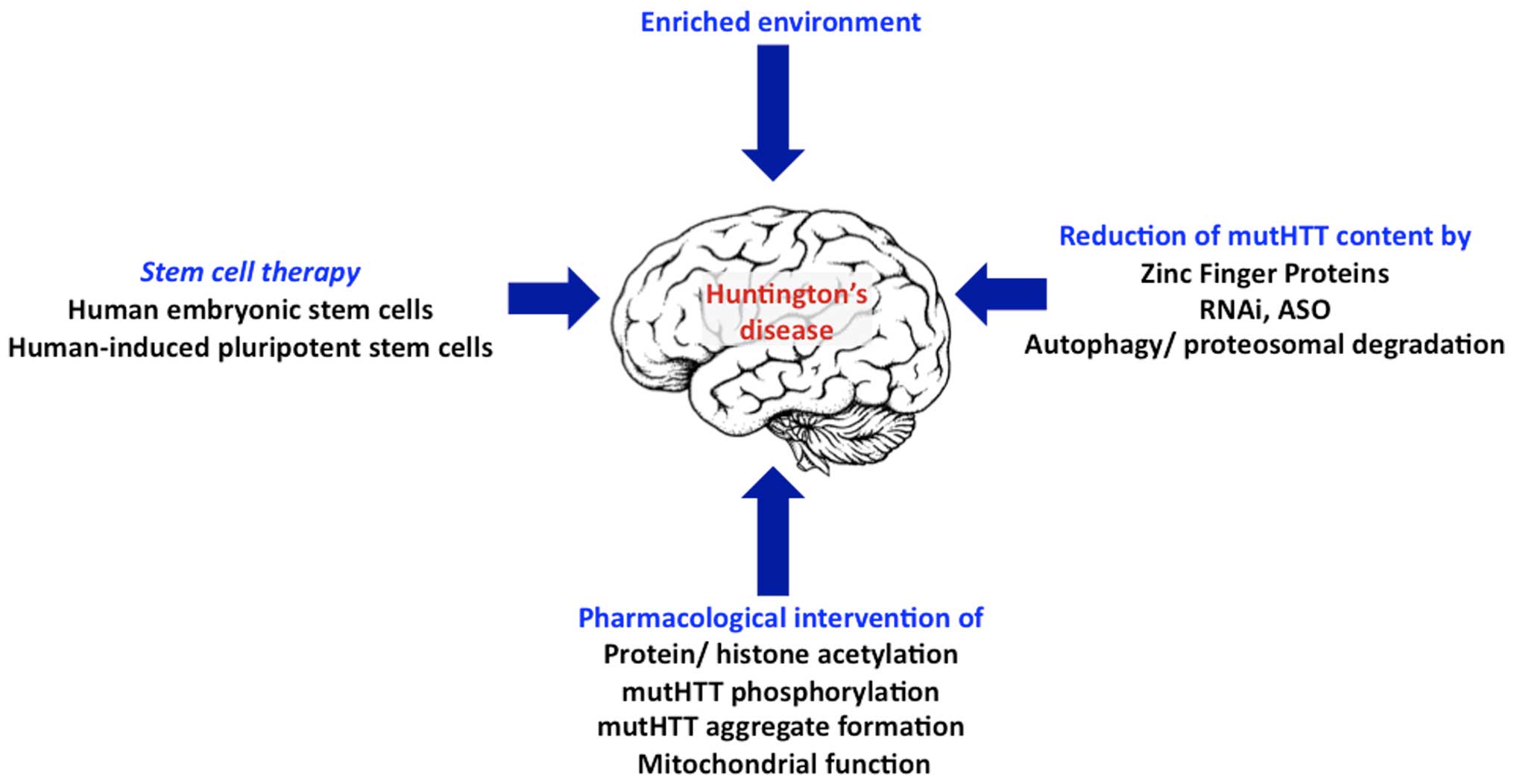
|
1
|
Ross CA, Aylward EH, Wild EJ, Langbehn DR,
Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS,
et al: Huntington disease: Natural history, biomarkers and
prospects for therapeutics. Nat Rev Neurol. 10:204–216. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pringsheim T, Wiltshire K, Day L, Dykeman
J, Steeves T and Jette N: The incidence and prevalence of
Huntington's disease: A systematic review and meta-analysis. Mov
Disord. 27:1083–1091. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bates G, Harper P and Jones L:
Huntington's disease. Oxford University Press; New York, NY:
2002
|
|
4
|
MacDonald M: The Huntington's Disease
Collaborative Research Group: A novel gene containing a
trinucleotide repeat that is expanded and unstable on Huntington's
disease chromosomes. Cell. 72:971–983. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Genetic Modifiers of Huntington's Disease
(GeM-HD) Consortium, . Identification of genetic factors that
modify clinical onset of Huntington's disease. Cell. 162:516–526.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graveland GA, Williams RS and DiFiglia M:
Evidence for degenerative and regenerative changes in neostriatal
spiny neurons in Huntington's disease. Science. 227:770–773. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mann DM, Oliver R and Snowden JS: The
topographic distribution of brain atrophy in Huntington's disease
and progressive supranuclear palsy. Acta Neuropathol. 85:553–559.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosas HD, Koroshetz WJ, Chen YI, Skeuse C,
Vangel M, Cudkowicz ME, Caplan K, Marek K, Seidman LJ, Makris N, et
al: Evidence for more widespread cerebral pathology in early HD: An
MRI-based morphometric analysis. Neurology. 60:1615–1620. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kemp JM and Powell TP: The structure of
the caudate nucleus of the cat: Light and electron microscopy.
Philos Trans R Soc Lond B Biol Sci. 262:383–401. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parent A, Bouchard C and Smith Y: The
striatopallidal and striatonigral projections: Two distinct fiber
systems in primate. Brain Res. 303:385–390. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heinsen H, Strik M, Bauer M, Luther K,
Ulmar G, Gangnus D, Jungkunz G, Eisenmenger W and Götz M: Cortical
and striatal neurone number in Huntington's disease. Acta
Neuropathol. 88:320–333. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vonsattel JP and DiFiglia M: Huntington
disease. J Neuropathol Exp Neurol. 57:369–384. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Myers RH: Huntington's disease genetics.
NeuroRx. 1:255–262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duyao M, Ambrose C, Myers R, Novelletto A,
Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M,
et al: Trinucleotide repeat length instability and age of onset in
Huntington's disease. Nat Genet. 4:387–392. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trottier Y, Biancalana V and Mandel JL:
Instability of CAG repeats in Huntington's disease: Relation to
parental transmission and age of onset. J Med Genet. 31:377–382.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Telenius H, Kremer B, Goldberg YP,
Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ,
Clarke LA, et al: Somatic and gonadal mosaicism of the Huntington
disease gene CAG repeat in brain and sperm. Nat Genet. 6:409–414.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zielonka D, Piotrowska I and Mielcarek M:
Cardiac dysfunction in huntington's disease. Exp Clin Cardiol.
20:2547–2554. 2014.
|
|
18
|
Zielonka D, Piotrowska I, Marcinkowski JT
and Mielcarek M: Skeletal muscle pathology in Huntington's disease.
Front Physiol. 5:3802014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Imarisio S, Carmichael J, Korolchuk V,
Chen CW, Saiki S, Rose C, Krishna G, Davies JE, Ttofi E, Underwood
BR, et al: Huntington's disease: From pathology and genetics to
potential therapies. Biochem J. 412:191–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Valor LM: Transcription, epigenetics and
ameliorative strategies in Huntington's Disease: A genome-wide
perspective. Mol Neurobiol. 51:406–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kennedy L, Evans E, Chen CM, Craven L,
Detloff PJ, Ennis M and Shelbourne PF: Dramatic tissue-specific
mutation length increases are an early molecular event in
Huntington disease pathogenesis. Hum Mol Genet. 12:3359–3367. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bečanović K, Nørremølle A, Neal SJ, Kay C,
Collins JA, Arenillas D, Lilja T, Gaudenzi G, Manoharan S, Doty CN,
et al: REGISTRY Investigators of the European Huntington's Disease
Network: A SNP in the HTT promoter alters NF-κB binding and is a
bidirectional genetic modifier of Huntington disease. Nat Neurosci.
18:807–816. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nasir J, Floresco SB, O'Kusky JR, Diewert
VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG and
Hayden MR: Targeted disruption of the Huntington's disease gene
results in embryonic lethality and behavioral and morphological
changes in heterozygotes. Cell. 81:811–823. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zeitlin S, Liu JP, Chapman DL, Papaioannou
VE and Efstratiadis A: Increased apoptosis and early embryonic
lethality in mice nullizygous for the Huntington's disease gene
homologue. Nat Genet. 11:155–163. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dragatsis I, Levine MS and Zeitlin S:
Inactivation of Hdh in the brain and testis results in progressive
neurodegeneration and sterility in mice. Nat Genet. 26:300–306.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hoffner G, Kahlem P and Djian P:
Perinuclear localization of huntingtin as a consequence of its
binding to microtubules through an interaction with beta-tubulin:
Relevance to Huntington's disease. J Cell Sci. 115:941–948.
2002.PubMed/NCBI
|
|
27
|
Godin JD, Colombo K, MolinaCalavita M,
Keryer G, Zala D, Charrin BC, Dietrich P, Volvert ML, Guillemot F,
Dragatsis I, et al: Huntingtin is required for mitotic spindle
orientation and mammalian neurogenesis. Neuron. 67:392–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
DiFiglia M, SenaEsteves M, Chase K, Sapp
E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et
al: Therapeutic silencing of mutant huntingtin with siRNA
attenuates striatal and cortical neuropathology and behavioral
deficits. Proc Natl Acad Sci USA. 104:17204–17209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arrasate M and Finkbeiner S: Protein
aggregates in Huntington's disease. Exp Neurol. 238:1–11. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ehrnhoefer DE, Sutton L and Hayden MR:
Small changes, big impact: Posttranslational modifications and
function of huntingtin in Huntington disease. Neuroscientist.
17:475–492. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zielonka D, Mielcarek M and Landwehrmeyer
GB: Update on Huntington's disease: Advances in care and emerging
therapeutic options. Parkinsonism Relat Disord. 21:169–178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeong H, Then F, Melia TJ Jr, Mazzulli JR,
Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al:
Acetylation targets mutant huntingtin to autophagosomes for
degradation. Cell. 137:60–72. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong G, Callegari E, Gloeckner CJ, Ueffing
M and Wang H: Mass spectrometric identification of novel
posttranslational modification sites in Huntingtin. Proteomics.
12:2060–2064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu X, Greiner ER, Mishra R, Kodali R,
Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R and Yang
XW: Serines 13 and 16 are critical determinants of full-length
human mutant huntingtin induced disease pathogenesis in HD mice.
Neuron. 64:828–840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zala D, Colin E, Rangone H, Liot G,
Humbert S and Saudou F: Phosphorylation of mutant huntingtin at
S421 restores anterograde and retrograde transport in neurons. Hum
Mol Genet. 17:3837–3846. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Venuto CS, McGarry A, Ma Q and Kieburtz K:
Pharmacologic approaches to the treatment of Huntington's disease.
Mov Disord. 27:31–41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mielcarek M, Benn CL, Franklin SA, Smith
DL, Woodman B, Marks PA and Bates GP: SAHA decreases HDAC 2 and 4
levels in vivo and improves molecular phenotypes in the R6/2 mouse
model of Huntington's disease. PLoS One. 6:e277462011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ravikumar B, Vacher C, Berger Z, Davies
JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, et
al: Inhibition of mTOR induces autophagy and reduces toxicity of
polyglutamine expansions in fly and mouse models of Huntington
disease. Nat Genet. 36:585–595. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Smith MR, Syed A, Lukacsovich T, Purcell
J, Barbaro BA, Worthge SA, Wei SR, Pollio G, Magnoni L, Scali C, et
al: A potent and selective Sirtuin 1 inhibitor alleviates pathology
in multiple animal and cell models of Huntington's disease. Hum Mol
Genet. 23:2995–3007. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Reilmann R, Squitieri F, Priller J, Saft
C, Mariotti C, Suessmuth S, Nemeth A, Tabrizi S, Quarrell O,
Craufurd D, et al: Safety and tolerability of selisistat for the
treatment of huntington's disease: Results from a randomized,
double-blind, placebo-controlled phase II trial. Neurology. (Suppl
10)82:S47.0042014.
|
|
41
|
Labbadia J and Morimoto RI: Huntington's
disease: Underlying molecular mechanisms and emerging concepts.
Trends Biochem Sci. 38:378–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Renna M, JimenezSanchez M, Sarkar S and
Rubinsztein DC: Chemical inducers of autophagy that enhance the
clearance of mutant proteins in neurodegenerative diseases. J Biol
Chem. 285:11061–11067. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Atwal RS, Desmond CR, Caron N, Maiuri T,
Xia J, Sipione S and Truant R: Kinase inhibitors modulate
huntingtin cell localization and toxicity. Nat Chem Biol.
7:453–460. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tam S, Geller R, Spiess C and Frydman J:
The chaperonin TRiC controls polyglutamine aggregation and toxicity
through subunit-specific interactions. Nat Cell Biol. 8:1155–1162.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sontag EM, Joachimiak LA, Tan Z, Tomlinson
A, Housman DE, Glabe CG, Potkin SG, Frydman J and Thompson LM:
Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates
mutant Huntingtin cellular phenotypes. Proc Natl Acad Sci USA.
110:3077–3082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gines S, Seong IS, Fossale E, Ivanova E,
Trettel F, Gusella JF, Wheeler VC, Persichetti F and MacDonald ME:
Specific progressive cAMP reduction implicates energy deficit in
presymptomatic Huntington's disease knock-in mice. Hum Mol Genet.
12:497–508. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sugars KL, Brown R, Cook LJ, Swartz J and
Rubinsztein DC: Decreased cAMP response element-mediated
transcription: An early event in exon 1 and full-length cell models
of Huntington's disease that contributes to polyglutamine
pathogenesis. J Biol Chem. 279:4988–4999. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Coskran TM, Morton D, Menniti FS,
Adamowicz WO, Kleiman RJ, Ryan AM, Strick CA, Schmidt CJ and
Stephenson DT: Immunohistochemical localization of
phosphodiesterase 10A in multiple mammalian species. J Histochem
Cytochem. 54:1205–1213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kleiman RJ, Kimmel LH, Bove SE, Lanz TA,
Harms JF, Romegialli A, Miller KS, Willis A, des Etages S, Kuhn M,
et al: Chronic suppression of phosphodiesterase 10A alters striatal
expression of genes responsible for neurotransmitter synthesis,
neurotransmission, and signaling pathways implicated in
Huntington's disease. J Pharmacol Exp Ther. 336:64–76. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wild EJ and Tabrizi SJ: Targets for future
clinical trials in Huntington's disease: What's in the pipeline?
Mov Disord. 29:1434–1445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gianfriddo M, Melani A, Turchi D,
Giovannini MG and Pedata F: Adenosine and glutamate extracellular
concentrations and mitogen-activated protein kinases in the
striatum of Huntington transgenic mice. Selective antagonism of
adenosine A2A receptors reduces transmitter outflow. Neurobiol Dis.
17:77–88. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morfini GA, You YM, Pollema SL, Kaminska
A, Liu K, Yoshioka K, Björkblom B, Coffey ET, Bagnato C, Han D, et
al: Pathogenic huntingtin inhibits fast axonal transport by
activating JNK3 and phosphorylating kinesin. Nat Neurosci.
12:864–871. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fan J, Gladding CM, Wang L, Zhang LY,
Kaufman AM, Milnerwood AJ and Raymond LA: P38 MAPK is involved in
enhanced NMDA receptor-dependent excitotoxicity in YAC transgenic
mouse model of Huntington disease. Neurobiol Dis. 45:999–1009.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Taylor DM, Moser R, Régulier E, Breuillaud
L, Dixon M, Beesen AA, Elliston L, Silva Santos MF, Kim J, Jones L,
et al: MAP kinase phosphatase 1 (MKP-1/DUSP1) is neuroprotective in
Huntington's disease via additive effects of JNK and p38
inhibition. J Neurosci. 33:2313–2325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Apostol BL, Simmons DA, Zuccato C, Illes
K, Pallos J, Casale M, Conforti P, Ramos C, Roarke M, Kathuria S,
et al: CEP-1347 reduces mutant huntingtin-associated neurotoxicity
and restores BDNF levels in R6/2 mice. Mol Cell Neurosci. 39:8–20.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mrzljak L: A05 targeting kmo: Basic
understanding and gaps. J Neurol Neurosurg Psychiatry. 85:A22014.
View Article : Google Scholar
|
|
57
|
Papworth M, Kolasinska P and Minczuk M:
Designer zinc-finger proteins and their applications. Gene.
366:27–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang H, Sun YM, Hao Y, Yan YP, Chen K,
Xin SH, Tang YP, Li XH, Jun T, Chen YY, Liu ZJ, Wang CR, Li H, Pei
Z, Shang HF, Zhang BR, Gu WH, Wu ZY, Tang BS and Burgunder JM:
Huntingtin gene CAG repeat numbers in Chinese patients with
Huntington's disease and controls. Eur J Neurol. 21:637–642. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
GarrigaCanut M, Agustín-Pavón C, Herrmann
F, Sánchez A, Dierssen M, Fillat C and Isalan M: Synthetic zinc
finger repressors reduce mutant huntingtin expression in the brain
of R6/2 mice. Proc Natl Acad Sci USA. 109:E3136–E3145. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zeitler J, Pearl JR, Froelich S, Yu Q,
Paschon DE, Miller JC, Marlen K, Guschin D, Narayanan A, Zhang L,
et al: Allele- specific repression of mutant Huntingtin expression
by engineered zinc finger transcriptional repressors as a potential
therapy for Huntington's disease. PNAS. 108:7052–7057.
2011.PubMed/NCBI
|
|
61
|
Li H, Haurigot V, Doyon Y, Li T, Wong SY,
Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al: In
vivo genome editing restores haemostasis in a mouse model of
haemophilia. Nature. 475:217–221. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kordasiewicz HB, Stanek LM, Wancewicz EV,
Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH,
Shihabuddin LS, et al: Sustained therapeutic reversal of
Huntington's disease by transient repression of huntingtin
synthesis. Neuron. 74:1031–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu XH and Yang XW: ‘Huntingtin holiday’:
Progress toward an antisense therapy for Huntington's disease.
Neuron. 74:964–966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Carroll JB, Warby SC, Southwell AL, Doty
CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM and Hayden
MR: Potent and selective antisense oligonucleotides targeting
single-nucleotide polymorphisms in the Huntington disease
gene/allele-specific silencing of mutant huntingtin. Mol Ther.
19:2178–2185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Miyagishi M, Hayashi M and Taira K:
Comparison of the suppressive effects of antisense oligonucleotides
and siRNAs directed against the same targets in mammalian cells.
Antisense Nucleic Acid Drug Dev. 13:1–7. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen ZJ, Kren BT, Wong PY, Low WC and
Steer CJ: Sleeping Beauty-mediated down-regulation of huntingtin
expression by RNA interference. Biochem Biophys Res Commun.
329:646–652. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Harper SQ, Staber PD, He X, Eliason SL,
Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL and Davidson BL:
RNA interference improves motor and neuropathological abnormalities
in a Huntington's disease mouse model. Proc Natl Acad Sci USA.
102:5820–5825. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Godinho BM, Malhotra M, O'Driscoll CM and
Cryan JF: Delivering a disease-modifying treatment for Huntington's
disease. Drug Discov Today. 20:50–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Drouet V, Perrin V, Hassig R, Dufour N,
Auregan G, Alves S, Bonvento G, Brouillet E, LuthiCarter R,
Hantraye P, et al: Sustained effects of nonallele-specific
Huntingtin silencing. Ann Neurol. 65:276–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Golas MM and Sander B: Use of human stem
cells in Huntington disease modeling and translational research.
Exp Neurol. 278:76–90. 2016. View Article : Google Scholar : PubMed/NCBI
|