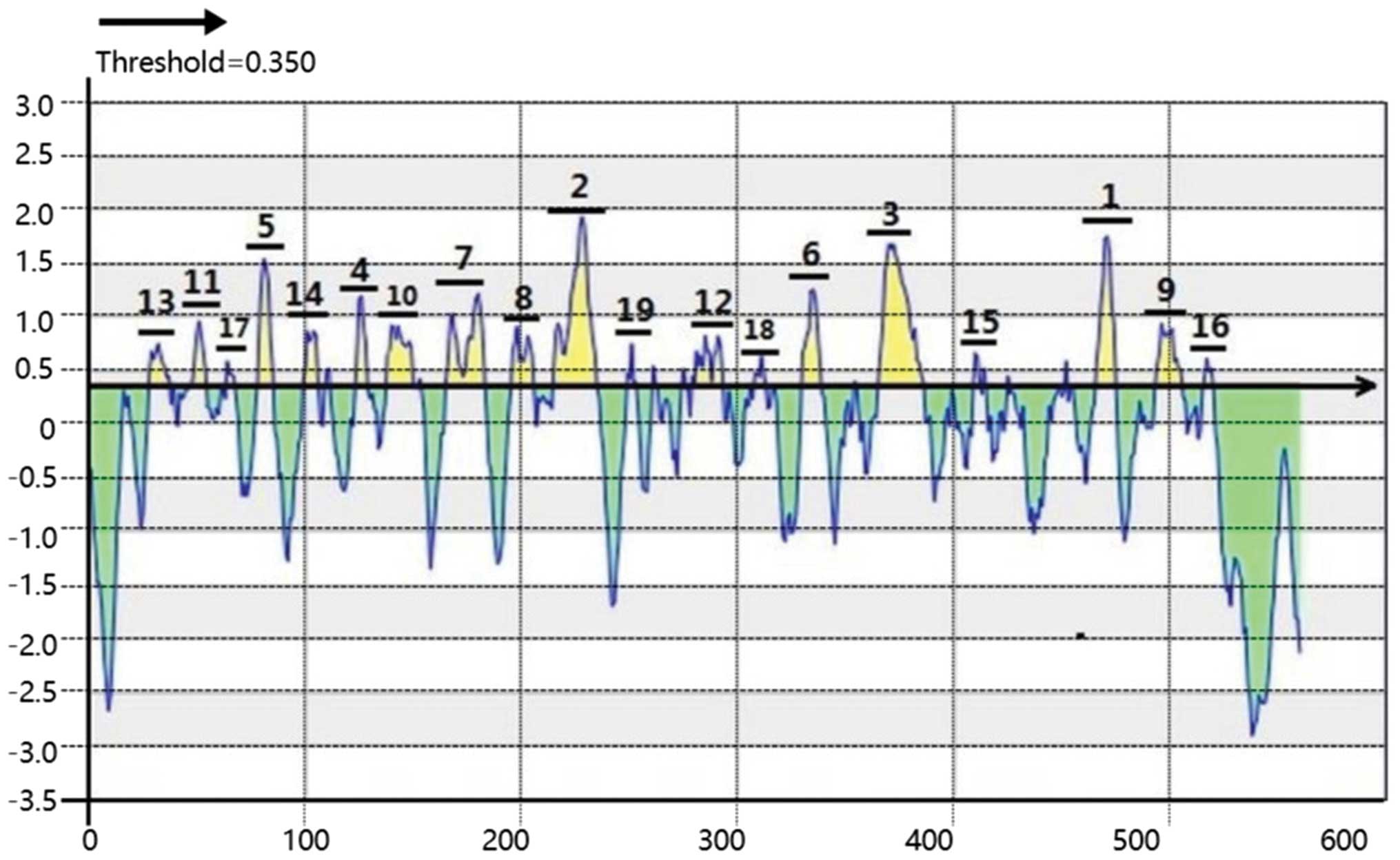
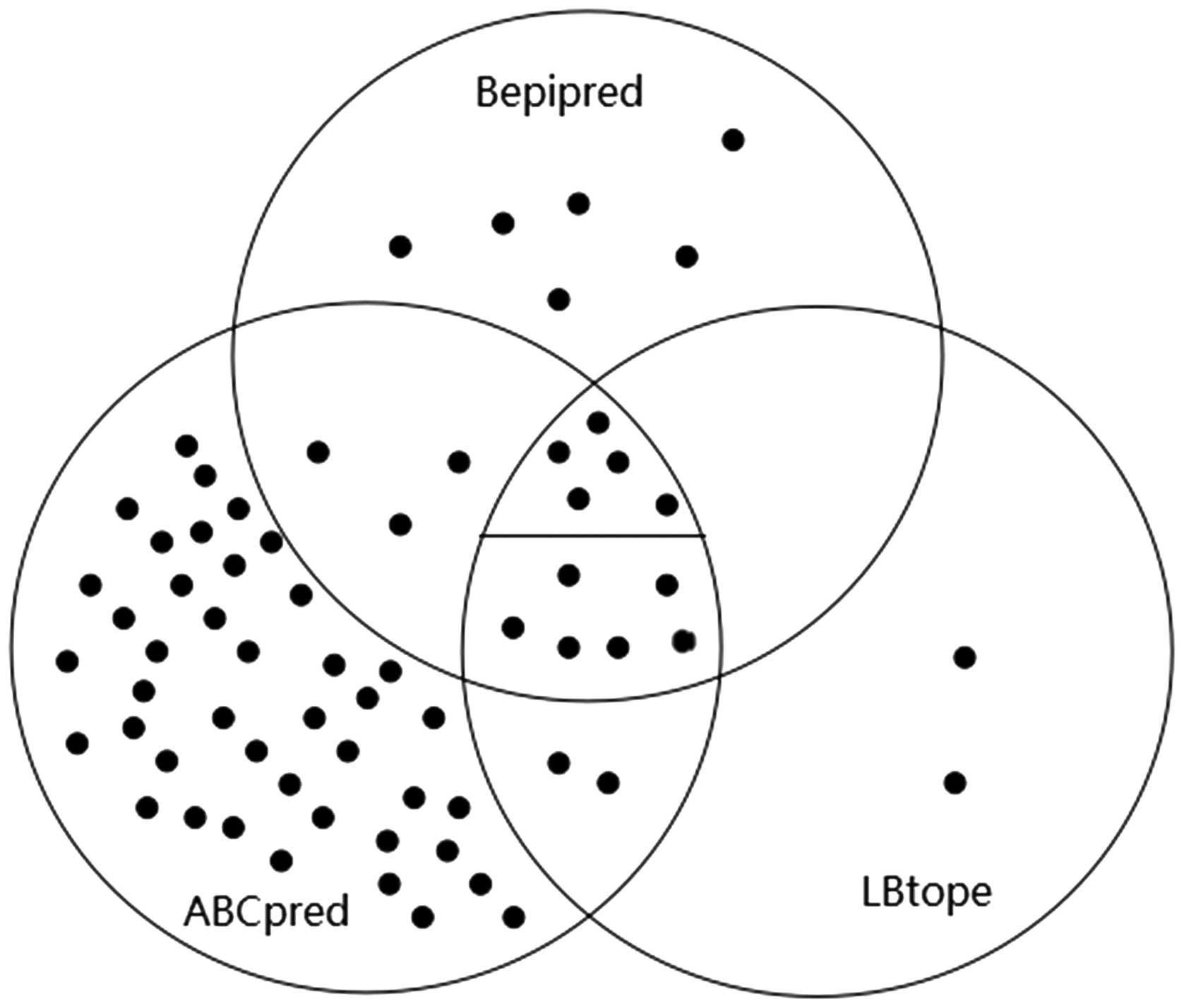
|
1
|
Klimov A, Prösch S, Schäfer J and Bucher
D: Subtype H7 influenza viruses: Comparative antigenic and
molecular analysis of the HA-, M-, and NS-genes. Arch Virol.
122:143–161. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fouchier RA, Schneeberger PM, Rozendaal
FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF,
Schutten M, Van Doornum GJ, et al: Avian influenza A virus (H7N7)
associated with human conjunctivitis and a fatal case of acute
respiratory distress syndrome. Proc Natl Acad Sci USA.
101:1356–1361. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
World Health Organization, . Human
infection with avian influenza A (H7N9) virus - update. http://www.who.int/csr/don/2014_01_20/en/May
1–2014
|
|
4
|
Sun Y, Shen Y and Lu H: Discovery process,
clinical characteristics, and treatment of patients infected with
avian influenza virus (H7N9) in Shanghai. Chin Med J (Engl).
127:185–186. 2014.PubMed/NCBI
|
|
5
|
Li YJ, Jiao XA, Pan ZM, Sun L, Wang CB,
Zhang SH, Sun QY and Liu XF: Development and characterization of
monoclonal antibodies against H7 hemagglutinin of avian influenza
virus. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23:953–955. 2007.(In
Chinese). PubMed/NCBI
|
|
6
|
Zhao B, Zhang X, Zhu W, Teng Z, Yu X, Gao
Y, Wu D, Pei E, Yuan Z, Yang L, et al: Novel avian influenza
A(H7N9) virus in tree sparrow, Shanghai, China, 2013. Emerg Infect
Dis. 20:850–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Russell RJ, Kerry PS, Stevens DJ,
Steinhauer DA, Martin SR, Gamblin SJ and Skehel JJ: Structure of
influenza hemagglutinin in complex with an inhibitor of membrane
fusion. Proc Natl Acad Sci USA. 105:17736–17741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Dai Z, Cheng H, Liu Z, Pan Z, Deng
W, Gao T, Li X, Yao Y, Ren J and Xue Y: Towards a better
understanding of the novel avian-origin H7N9 influenza A virus in
China. Sci Rep. 3:23182013.PubMed/NCBI
|
|
9
|
Barlow DJ, Edwards MS and Thornton JM:
Continuous and discontinuous protein antigenic determinants.
Nature. 322:747–748. 1986. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Evans MC: Recent advances in
immunoinformatics: Application of in silico tools to drug
development. Curr Opin Drug Discov Devel. 11:233–241.
2008.PubMed/NCBI
|
|
11
|
Van Regenmortel MH: Synthetic peptides
versus natural antigens in immunoassays. Ann Biol Clin (Paris).
51:39–41. 1993.PubMed/NCBI
|
|
12
|
Saha S and Raghava GP: Prediction of
continuous B-cell epitopes in an antigen using recurrent neural
network. Proteins. 65:40–48. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu WF, Gao RB, Wang DY, Yang L, Zhu Y and
Shu YL: A review of H7 subtype avain influenza virus. Bing Du Xue
Bao. 29:245–249. 2013.(In Chinese). PubMed/NCBI
|
|
14
|
Larsen JE, Lund O and Nielsen M: Improved
method for predicting linear B-cell epitopes. Immunome Res.
2:22006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parker JM, Guo D and Hodges RS: New
hydrophilicity scale derived from high-performance liquid
chromatography peptide retention data: Correlation of predicted
surface residues with antigenicity and X-ray-derived accessible
sites. Biochemistry. 25:5425–5432. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh H, Ansari HR and Raghava GP:
Improved method for linear B-cell epitope prediction using
antigen's primary sequence. PLoS One. 8:e622162013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu R, de Vries RP, Zhu X, Nycholat CM,
McBride R, Yu W, Paulson JC and Wilson IA: Preferential recognition
of avian-like receptors in human influenza A H7N9 viruses. Science.
342:1230–1235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krchnák V, Mach O and Malý A: Computer
prediction of B-cell determinants from protein amino acid sequences
based on incidence of beta turns. Methods Enzymol. 178:586–611.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pellequer JL, Westhof E and Van
Regenmortel MH: Correlation between the location of antigenic sites
and the prediction of turns in proteins. Immunol Lett. 36:83–99.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dudek NL, Perlmutter P, Aguilar MI, Croft
NP and Purcell AW: Epitope discovery and their use in peptide based
vaccines. Curr Pharm Des. 16:3149–3157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bryson CJ, Jones TD and Baker MP:
Prediction of immunogenicity of therapeutic proteins: Validity of
computational tools. BioDrugs. 24:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Groot AS, Sbai H, Aubin CS, McMurry J
and Martin W: Immuno-informatics: Mining genomes for vaccine
components. Immunol Cell Biol. 80:255–269. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frikha-Gargouri O, Gdoura R, Znazen A,
Gargouri B, Gargouri J, Rebai A and Hammami A: Evaluation of an in
silico predicted specific and immunogenic antigen from the OmcB
protein for the serodiagnosis of Chlamydia trachomatis infections.
BMC Microbiol. 8:2172008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jones MS and Carter JM: Prediction of
B-cell epitopes in listeriolysin O, a cholesterol dependent
cytolysin secreted by listeria monocytogenes. Adv Bioinformatics.
2014:8716762014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maksimov P, Zerweck J, Maksimov A, Hotop
A, Gross U, Pleyer U, Spekker K, Däubener W, Werdermann S,
Niederstrasser O, et al: Peptide microarray analysis of in
silico-predicted epitopes for serological diagnosis of Toxoplasma
gondii infection in humans. Clin Vaccine Immunol. 19:865–874. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saha S, Bhasin M and Raghava GP: Bcipep: A
database of B-cell epitopes. BMC Genomics. 6:792005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karplus PA and Schulz GE: Prediction of
chain flexibility in proteins. Naturwissenschaften. 72:212–213.
1985. View Article : Google Scholar
|
|
28
|
Emini EA, Hughes JV, Perlow DS and Boger
J: Induction of hepatitis A virus-neutralizing antibody by a
virus-specific synthetic peptide. J Virol. 55:836–839.
1985.PubMed/NCBI
|
|
29
|
Costa JG, Faccendini PL, Sferco SJ, Lagier
CM and Marcipar IS: Evaluation and comparison of the ability of
online available prediction programs to predict true linear B-cell
epitopes. Protein Pept Lett. 20:724–730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reimer U: Prediction of linear B-cell
epitopes. Methods Mol Biol. 524:335–344. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kageyama T, Fujisaki S, Takashita E, Xu H,
Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y and Tashiro M:
Genetic analysis of novel avian A (H7N9) influenza viruses isolated
from patients in China, February to April 2013. Euro Surveill.
18:204532013.PubMed/NCBI
|
|
32
|
Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W,
Bi Y, Wu Y, Li X, Yan J, et al: Origin and diversity of novel avian
influenza A H7N9 viruses causing human infection: Phylogenetic,
structural, and coalescent analyses. Lancet. 381:1926–1932. 2013.
View Article : Google Scholar : PubMed/NCBI
|