Introduction
The overall incidence of gastric cancer has
decreased in a number of countries as a result of extensive
diagnostic and therapeutic investigations. However, gastric cancer
remains a major global health burden and is the second most common
cause of cancer-related mortality (1). The geographic distribution of gastric
cancer is characterized by wide international variations,
particularly in high-risk areas such as Japan, Korea, China, and
South and Central America (2).
To date, molecular and genetic abnormalities are
responsible for the development and progression of gastric cancer,
including the activation of oncogenes and inactivation of various
tumor suppressor genes (3,4). As a result of gene alteration, the
downstream signal transduction pathways will be consequently
affected, involving the control of diverse cellular functions such
as cell growth, differentiation, metastasis and adhesion (5,6). Wang
et al (7) indicated that
specificity protein 1 (Sp1) was significantly increased in gastric
tumor specimens and associated with patient survival, suggesting
that abnormal Sp1 expression contributes towards the development
and progression of gastric cancer. Kurayoshi et al (8) proposed that the overexpression of Wnt
family member 5A improved the migration and invasion ability of
gastric cancer cells and was associated with the aggressiveness and
poor prognosis of gastric cancer. Further evidence demonstrated
that the restoration of Kruppel-like factor 4 expression resulted
in the marked suppression of gastric cancer cell growth in
vitro and significant attenuation of tumor growth in an animal
model, suggesting that it may serve as a prognostic marker and
potential therapeutic target for gastric cancer (9).
Peptidylarginine deiminase type 4 (PADI4),
one of four known PADI genes, is characterized by the conversion of
peptidylarginine into citrulline in the process of citrullination.
The gene is expressed in hematopoietic progenitor cells,
granulocytes, T cells, B cells, macrophages and natural killer
cells (10). Accumulating evidence
suggests that PADI4 appears to be overexpressed in numerous types
of carcinomas, including breast carcinoma, hepatocarcinoma, renal,
bladder and lung carcinoma (11).
Chang et al (12) indicated
that the expression levels of PADI4 mRNA and protein are
significantly enhanced in breast fibroadenoma and thyroid adenoma
compared with surrounding healthy tissues. In addition, one study
suggested that PADI4 was expressed at higher levels in ovarian
adenocarcinoma, and confirmed this using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses (13). Ordóñez
et al (14) suggested that
PADI4 may have an effect on tumor progression with regards to the
elevation of citrullinated antithrombin levels in serum samples of
colorectal adenocarcinoma patients. On this basis, the detection of
genetic abnormalities of PADI4 may provide an alternative
opportunity for the early diagnosis and clinical interventions of
various cancers.
In the present study, the expression of PADI4 was
detected in gastric cancer and normal gastric mucosa tissues. PADI4
was suppressed with the aid of small interfering RNA (siRNA) and
5-fluorouracil (5-Fu), and the effects of silencing PADI4 on
various cell functions of SGC-7901 and AGS cells were determined in
order to explore the pathogenic role of PADI4 in gastric
cancer.
Materials and methods
Materials and reagents
A total of 10 tissues samples from patients with
gastric cancer (6 men and 4 women) between February 2010 and
October 2012 were obtained at the Third People's Hospital of
Qingdao (Qingdao, China). Ten normal gastric mucosa tissues (a
distance away from the resected margin of gastric cancer) were
obtained from these patients to use as a control. All samples were
confirmed by pathological analysis and were not treated with
radiotherapy and chemotherapy prior to surgery. The median age in
the cohort of patients was 55.3±5.9 years (range, 47.6–57.6 years).
All samples were stored at −80°C for further analysis. Written
informed consent was obtained from all the patients. The study was
conducted with approval from the Ethics Committee of the Third
People's Hospital of Qingdao.
Roswell Park Memorial Institute (RPMI)-160 culture
medium, fetal bovine serum (FBS) and double antibody
(penicillin-streptomycin) were obtained from Gibco (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). PADI4 and GAPDH sheep
anti-rabbit antibodies, and secondary antibody, were obtained from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Radioimmunoprecipitation assay (RIPA) lysis buffer, polyvinylidene
difluoride (PVDF) membrane and Bradford Protein Quantitative kit
were purchased from Beyotime Institute of Biotechnology (Jiangsu,
China). Enhanced Chemiluminescence (ECL) kit was obtained from
Invitrogen (Thermo Fisher Scientific, Inc.). Bovine serum albumin
(BSA) was obtained from Sangon Biotechnology Co., Ltd. (Shanghai,
China). Total RNA Extraction kit was obtained from Omega Bio-tek,
Inc. (Doraville, GA, USA). Quantitative PCR kit and SYBR Green qPCR
Master Mix was obtained from Hoffmann-La Roche, Inc. (Basel,
Switzerland). 5-Fu was obtained from Anji Haosen Pharmaceutical
Co., Ltd. (Jiangsu, China).
Cell culture and treatment with siRNA
and 5-Fu
Human gastric cancer cell lines (SGC-7901 and AGS)
were cultured in RPMI-1640 supplemented with 10% FBS at 37°C with
5% CO2 in a humidified atmosphere. Cells in logarithmic
growth phase were resuspended and seeded in a 6-well plate at a
density of 5×105 cells/ml. Subsequently, transient
transfection was performed when cells reached 80% confluence. For
the experimental transfection, samples were divided into the
following groups: Mock group (subjected to transfection reagent);
negative group (subjected to siRNA transfection); PADI4 siRNA group
(subjected to PADI4 siRNA transfection); 5-Fu group (subjected to
5-Fu); and 5-Fu+ siRNA transfection group (treated with
5-Fu and subjected to PADI4 siRNA transfection). All transfection
procedures were performed according to the manufacturer's
instructions of Lipofectamine 2000. The interference sequences in
the PADI4 siRNA and negative group were as follows: siPADI4 sense,
5′GAAGGAGUUUCCCAUCAAATT'3 and antisense, 5′UUUGAUGGGAAACUCCUUCAG'3;
negative control sense, UUCUCCGAACGUGUCACG and antisense,
5′ACGUGACACGUUCGGAGAATT'3.
Western blot analysis
Fresh tissues were cut into sections and lysed in
RIPA buffer on ice. Cell lysates were centrifuged at 12,000 rpm for
10 min, and the protein supernatant was transferred into new tubes.
Subsequently, the protein concentration of each sample was
determined using BCA Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.). For SDS-PAGE, 20 µg of the total protein from
each sample was resolved using 12% SDS-PAGE and transferred to PVDF
membranes for 45 min using a tank transfer system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Then, the membranes were
blocked in Tris-buffered saline containing Tween 20 (TBST) with 3%
BSA at 37°C for 1 h, and incubated with PADI4 primary antibody
(1:500; cat. no. sc-365369) overnight at 4°C. After washing three
times with TBST, the membranes were incubated with anti-rabbit
secondary antibody (1:500; cat. no. sc-98991) for 1 h at room
temperature. Proteins were detected using an ECL kit according to
the manufacturer's instructions. GAPDH was used as an internal
control.
RT-qPCR
Total RNA from the gastric tissues was extracted
with TRIzol reagent according to the manufacturer's instructions as
previously described (15). cDNA
synthesis was performed with ~500 ng RNA using the Superscript RT
kit (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR
amplification was performed using the Mx3000P QPCR System (Agilent
Technologies, Inc., Santa Clara, CA, USA) with the primers listed
in Table I. As an internal control
for qPCR, GAPDH mRNA expression was amplified from the same cDNA
samples. All results were normalized to GAPDH amplification. PCR
reactions were performed in a total volume of 20 µl, containing 15
µl 1X SYBR Green Supermix, 1 µl each specific primer, 2 µl water
and 1 µl cDNA template. The amplification reaction conditions were
as follows: Pre-denaturation at 90°C for 3 min; followed by 40
cycles of denaturation at 94°C for 30 sec; annealing at 30°C for 30
sec; and extension at 72°C for 40 sec. The 2−ΔΔCq method
was applied to analyze the relative changes in gene expression from
RT-qPCR experiments, as previously described (16).
 | Table I.Specific primers of PADI4, MMP2, MMP9
and GAPDH. |
Table I.
Specific primers of PADI4, MMP2, MMP9
and GAPDH.
| Gene | Forward primer | Reverse primer |
|---|
| PADI4 |
5′-GGGGACATTGATCCGTGTGA-3′ |
5′-TCGTCAGGGTCACCTCTACC-3′ |
| MMP2 |
5′-CCAGCTGGCCTAGTGATGAT-3′ |
5′-CCGCATGGTCTCGATGGTAT-3′ |
| MMP9 |
5′-TCTATGGTCCTCGCCCTGAA-3′ |
5′-CATCGTCCACCGGACTCAAA-3′ |
| GAPDH |
5′-AATGGGCAGCCGTTAGGAAA-3′ |
5′-GCGCCCAATACGACCAAATC-3′ |
MTT assay
An MTT assay was performed to determine the effect
of PADI4 on the cellular growth of SGC-7901 and AGS cells, as
previously described (17). Briefly,
SGC-7901 and AGS cells at a density of 5×103 cells/ml
were seeded in 96-well plates and cultured with the various
transfections or 5-Fu, as described above, for 24, 48 and 72 h.
Subsequently, 20 µl of MTT (5 mg/ml; Gefan Biotech, Shanghai China)
was added to each well and incubated for 4 h at 37°C. Then, 150 µl
dimethyl sulfoxide was added to each well, and the mixture was
shaken in a horizontal direction for 10 min to dissolve the
produced formazan crystals. The optical density (OD) value at 490
nm of each sample was measured using a plate reader.
Flow cytometry assay
Cell cycle was determined by flow cytometry, as
previously described (18). Briefly,
cells in the logarithmic growth phase were collected subsequent to
transfection with PADI4 siRNA for 48 h. Cells were fixed with 70%
ethanol for 24 h followed by washing in PBS three times.
Subsequently, cells were treated with 50 µl RNAse solution (0.5
mg/ml) at 37°C for 30 min. Then, cells were incubated with
propidium iodide (50 µg/ml) for staining at 4°C for 30 min under
dark conditions and filtered with a stainless steel screen (200
mesh). The cell cycle of each sample was detected using flow
cytometry. Experiments were repeated three times.
Cell invasion assay
Cell invasion was identified using a Transwell cell
culture chamber (8 µm; Corning Incorporated, Corning, NY, USA), as
previously described (19). Briefly,
SGC-7901 and AGS cells were suspended in serum-free RPMI-l640
culture medium at a concentration of 1×105 cells/ml
following transfections, as described above. Cell suspension (200
µl) was then added to the upper chamber (2×104
cells/well). Simultaneously, RPMI-1640 media supplemented with 20%
FBS was added to the lower compartment. Then, the Transwell chamber
was incubated at 37°C in a 5% CO2 atmosphere for 24 h.
Subsequently, cells in the upper compartment were collected and
fixed with formaldehyde solution. Following staining with
hematoxylin and eosin, five fields of each image were randomly
selected at higher magnification using a confocal microscope, and
the number of cells were calculated. Each assay was repeated three
times.
Statistical analysis
Statistical analyses were performed by one-way
analysis of variance using SPSS version 13.0 (SPSS, Inc., Chicago,
IL, USA). Data are presented as mean ± standard deviation.
Student's t-test was performed to evaluate inter-group comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of PADI4 in gastric cancer
and normal mucosa tissue
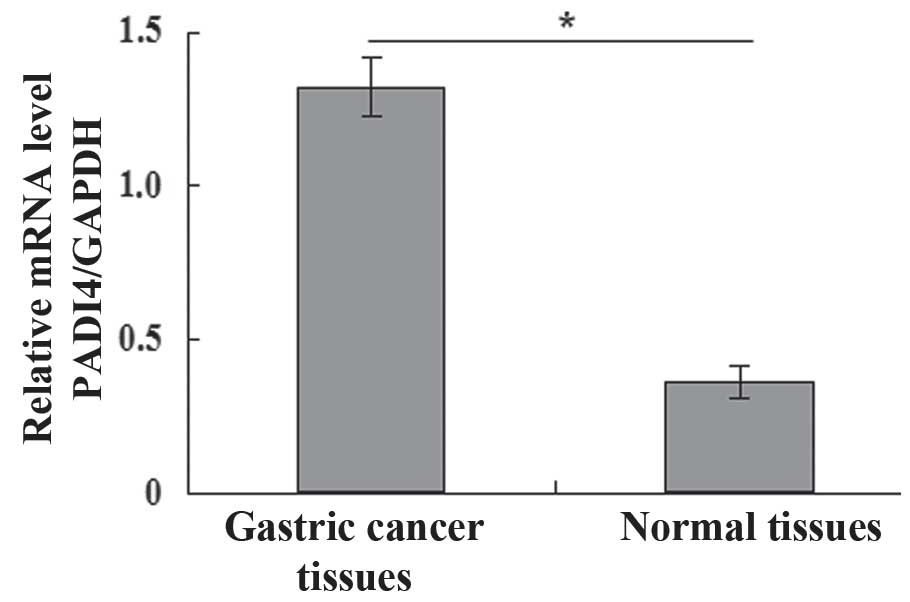
Fig. 1 presents the
expression levels of PADI4 in gastric cancer and normal mucosa
tissues. A significant increase in PADI4 protein mRNA expression
levels were identified in gastric cancer tissue compared with
normal mucosa tissue (P<0.05; Fig.
1). Furthermore, the expression of PADI4 (1.367±0.268) in
gastric cancer tissue was significantly higher compared with normal
mucosa tissue (0.429±0.335), analyzed by RT-qPCR analysis
(P<0.05).
Effects of silencing PADI4 on the
proliferation, apoptosis and invasion of SGC-7901 and AGS
cells
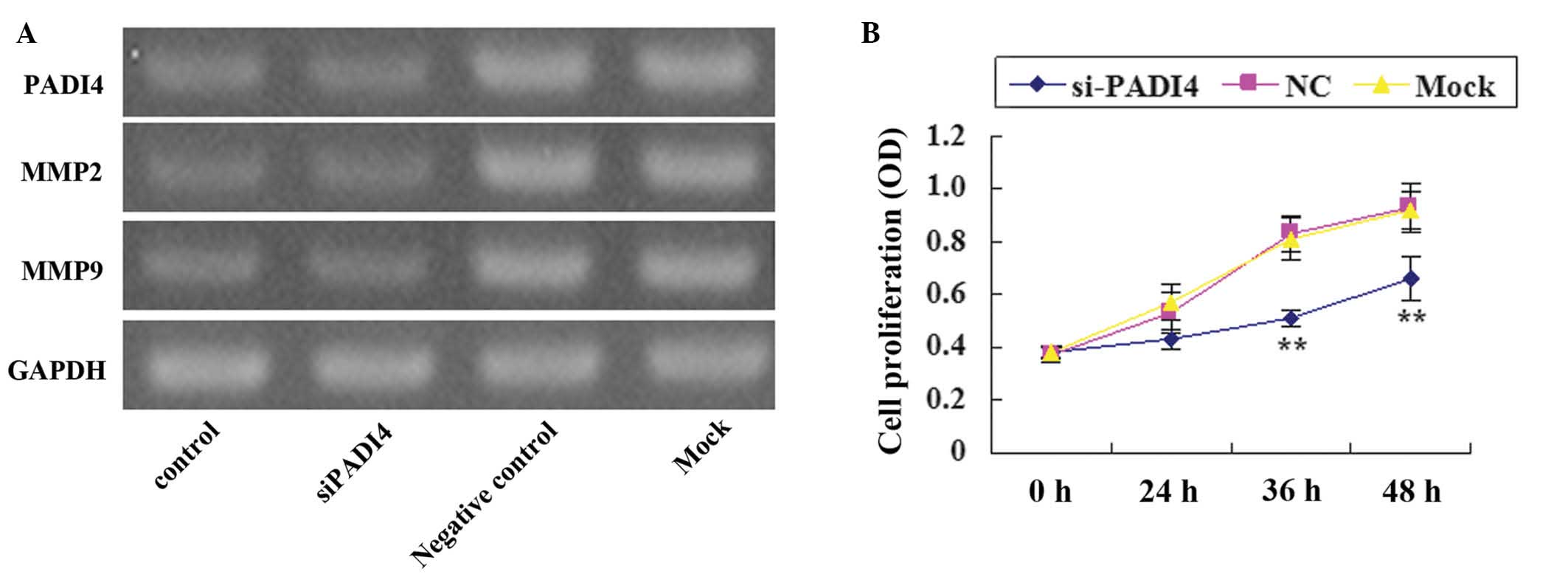
Following silencing PADI4 with siRNA, the expression
level of PADI4 was markedly decreased compared with the mock and
negative group (Fig. 2A). A
significant decrease (P<0.05) in the proliferation of SGC-7901
and AGS cells in the PADI4 siRNA group was identified in comparison
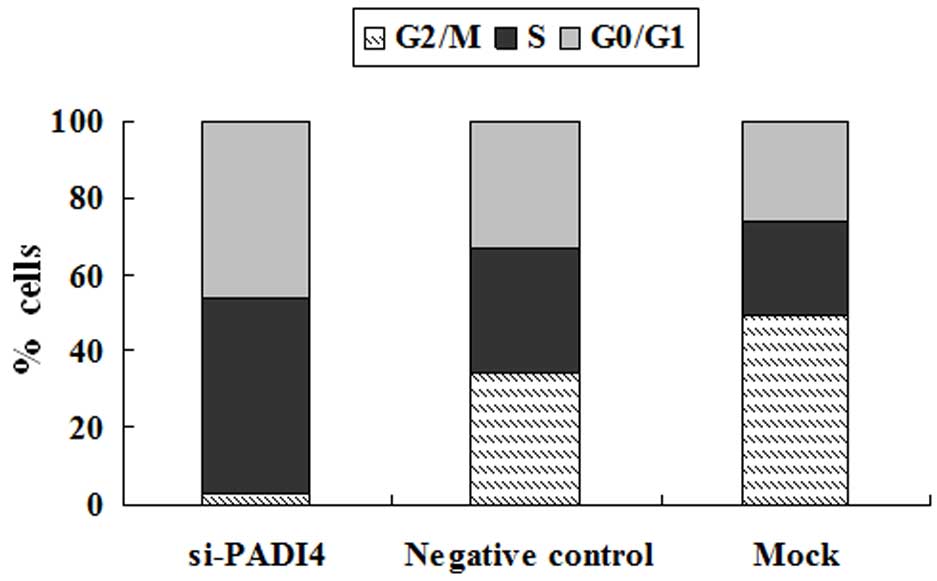
with the mock and negative groups at 36 and 48 h (Fig. 2B). In addition, marked S phase arrest
and a marked decrease in the number of gastric cancer cells in the
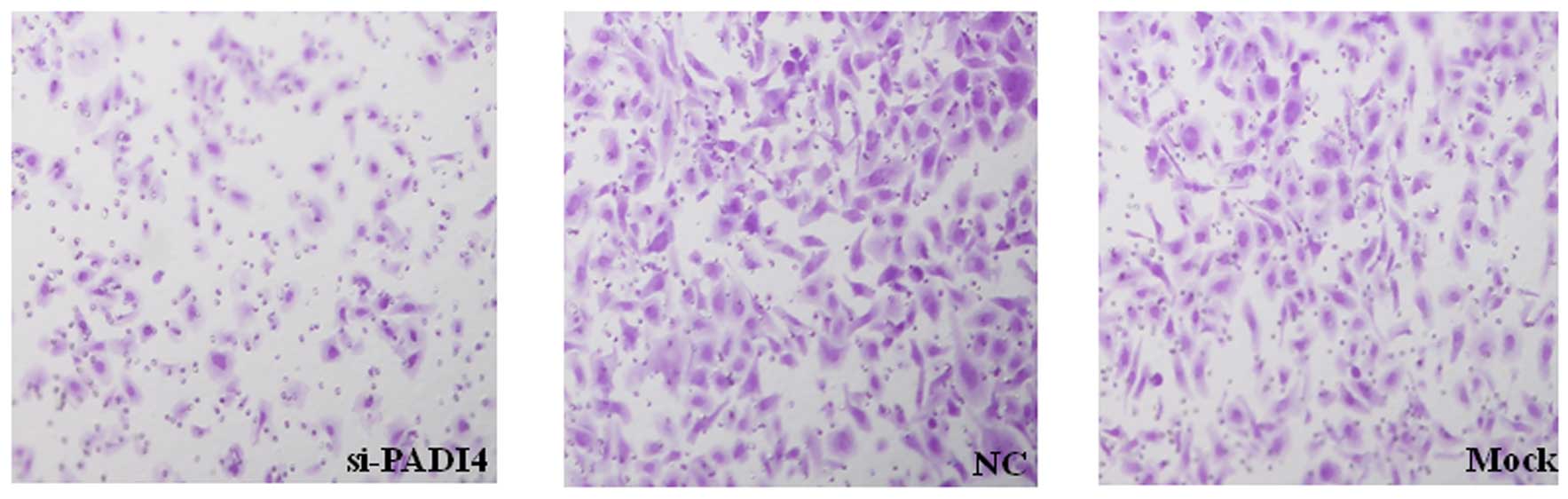
G2/M phase was identified in the PADI4 siRNA group (Fig. 3). Furthermore, the invasion of
SGC-7901 and AGS cells were markedly decreased when treated with
PADI4 siRNA (Fig. 4).
Effect of silencing PADI4 on the
expression of matrix metalloproteinase (MMP) 2 and MMP9 in gastric
cancer cells
After silencing PADI4 in SGC-7901 cells, MMP2 and
MMP9 expression levels were lower compared with the mock and
negative control groups (Fig.
2A).
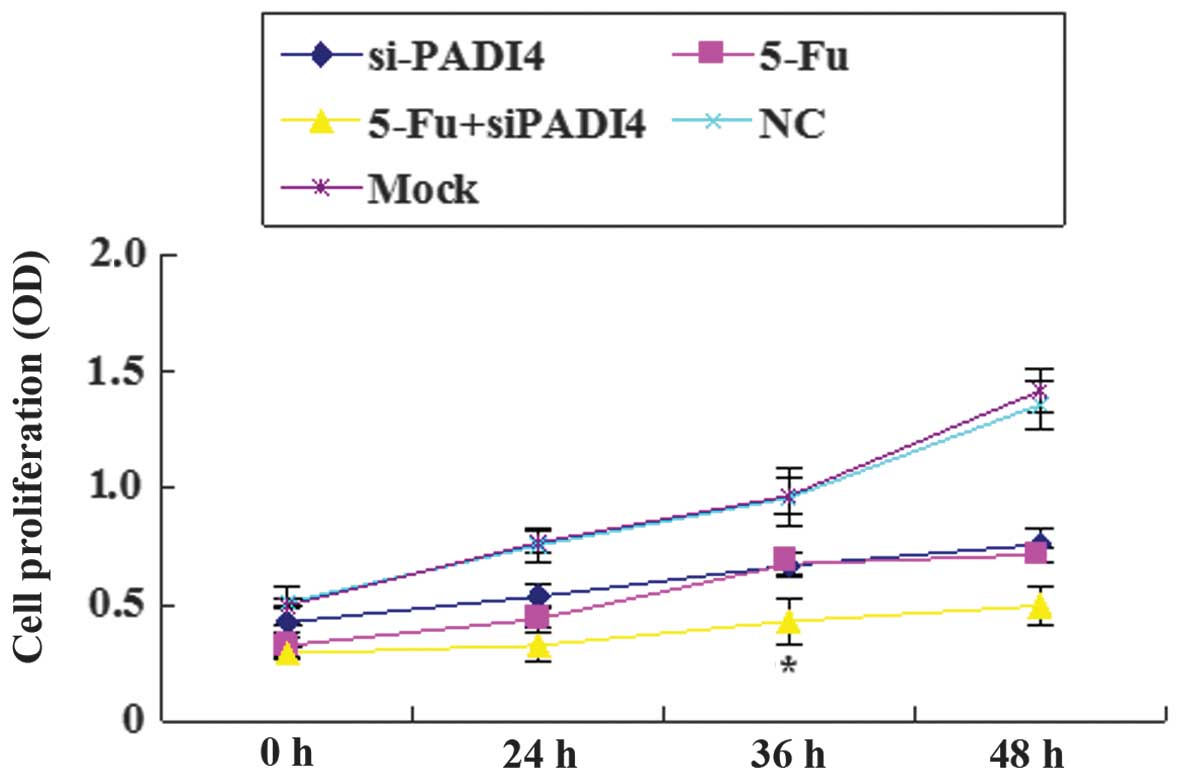
Synergistic effect of PADI4 siRNA and
5-Fu on the proliferation of SGC-7901 and AGS cells
A marked decrease was observed in the proliferation
of gastric cancer cells in the PADI4 siRNA, 5-Fu and PADI4 siRNA +
5-Fu groups compared with the mock and control groups. The
inhibitory effect was most prominent when PADI4 siRNA and 5-Fu were
combined. Furthermore, a significant difference (P<0.05) was
identified in the combination group at 36 h compared with the PADI4
siRNA group and the 5-Fu group (Fig.
5).
Discussion
PADI4 is one member of the PADI gene family
catalyzing protein cirtullination in the presence of
Ca2+. A number of reports have suggested that
citrullination is associated with specific biological events
involving apoptosis, inflammation, histone-related gene expression
and trauma (20–22). Extensive studies have focused on the
role of PADI4 in rheumatoid arthritis (RA). Evidence indicates that
PADI4 presents a strong association with RA by whole genome single
nucleotide polymorphism scanning (23). Chang et al (24) demonstrated an increased expression of
PADI4 in the synovial fluid and synovial membrane of patients with
RA. Iwamoto et al (25)
indicated that PADI was positively correlated with RA in Japanese
and Caucasian populations of European descent with a meta-analysis.
In addition, the role of PADI4 has attracted increasing attention
with regards to a number of malignant tumors. Lv et al
(26) observed a significant
elevation of PADI4 in hepatocellular carcinomas compared with
surrounding healthy tissue using western blot analysis. Ulivi et
al (27) demonstrated that
non-small-cell lung cancer (small-cell lung cancer) could be
accurately discriminated according to the expression of PADI4 and
pro-platelet basic protein by free circulating DNA analysis.
In the present study, a significant increase in
PADI4 mRNA expression levels was identified in gastric cancer
tissues compared with normal gastric mucosa tissues, suggesting
that there is a correlation between PADI4 and gastric cancer, and
that it may serve a role clinical diagnosis. In addition, it was
observed that the proliferation and invasion of SGC-7901 and AGS
cells were significantly decreased when PADI4 was silenced with
siRNA. Furthermore, silencing PADI4 resulted in significant S phase
arrest and a marked decrease in the number of cells in the G2/M
phase. Together, these results suggest that overexpression of PADI4
contributes towards gastric cancer cell growth and migration, and
that the silencing of PADI4 may provide an alternative treatment
for gastric cancer.
MMPs are a family of enzymes with proteolytic
activity for degrading various components of the extracellular
matrix (ECM). Among these are MMP2 and MMP9 (gelatinase subgroup of
MMPs) which serve an important role in the regulation of key
signaling pathways in cell invasion, growth, angiogenesis and
inflammation, by cleaving numerous different targets, such as
cytokines, chemokines, growth factors and ECM (28). Currently, growing research focuses on
MMP2 and MMP9 since they are overexpressed in various malignant
tumors, and their expression and activity are frequently related to
tumor progression and poor prognosis (29). Previous evidence suggests that the
expression of MMP2 and MMP9 increases in a numbers of carcinomas,
such as brain, ovarian, breast and lung cancers (30,31).
Wong et al (32) proposed
that MMP2 and MMP9 contribute towards the aggressiveness of highly
metastatic forms of nasopharyngeal carcinoma. Matsumura et
al (33) indicated that a MMP-9
polymorphism was associated with the depth of tumor invasion and
tumor, nodes and metastasis classification of gastric cancer. In
the current study, the expression of MMP2 and MMP9 was decreased
after silencing PADI4 in SGC-7901 and AGS cells, which suggests
that PADI4 may contribute towards the abnormal invasion of gastric
cancer cells by regulating the expression of MMP2 and MMP9.
Aside from the role of PADI4 in cell invasion, PADI4
serves a crucial role in mediating cell cycle and apoptosis, both
of which are associated with the expression of the tumor suppressor
p53. Liu et al (34)
demonstrated that overexpression of PADI4 could upregulate the
expression of p53 and its downstream factors p21 and B-cell
lymphoma 2-associated X protein, result in cell cycle arrest in the
G1 phase, and cause mitochondria-mediated apoptosis of human
leukemia HL-60 cells and human acute T leukemia Jurkat cells.
Further evidence indicated that the depletion of PADI4 by small
hairpin RNA increased the population of G1 cells, decreased the
population of S and G2/M cells and increased cell apoptosis in
HCT116 cells in a p53-dependent manner (35). Similar results were obtained in
another study, where Yao et al (36) demonstrated that knockdown of PADI4
with Cl-amidine and siRNA decreased cell viability and induced
apoptosis of breast cancer MCF-7 cells. In the present study, it
was observed that the silencing of PADI4 resulted in an S cell
cycle block with a concomitant decrease of cells in the G2/M phase,
and decreased cell growth and invasion. Furthermore, the inhibitory
effect of PADI4 siRNA on the proliferation of gastric cancer cells
were enhanced when combined with 5-Fu. Together, the results
suggest that PAD4 siRNA treatment coupled with 5-Fu may contribute
towards the decrease in drug resistance of cancer cells, improve
chemotherapeutic effects and provide a possible therapeutic option
for treating gastric cancer.
In conclusion, the present study demonstrates the
increased expression of PADI4 in gastric cancer tissues. The
silencing of PADI4 suppressed the proliferation and invasion of
cancer cells, and arrested cell cycle in the S phase. In addition,
the combination of siRNA PADI4 with 5-Fu presented a stronger
inhibitory effect on gastric cell growth compared with siRNA PADI4
alone. As a result, PADI4 may be considered as a prognostic
indicator of gastric tumor and a potential target for gastric
cancer therapy.
References
|
1
|
Roder DM: The epidemiology of gastric
cancer. Gastric Cancer. 5(Suppl 1): 5–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
1): S4–S66. 2001. View Article : Google Scholar
|
|
3
|
Tahara E: Molecular aspects of invasion
and metastasis of stomach cancer. Verh Dtsch Ges Pathol. 84:43–49.
2000.PubMed/NCBI
|
|
4
|
Sud R, Wells D, Talbot IC and Delhanty JD:
Genetic alterations in gastric cancers from British patients.
Cancer Genet Cytogenet. 126:111–119. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fiocca R, Luinetti O, Villani L, Mastracci
L, Quilici P, Grillo F and Ranzani GN: Molecular mechanisms
involved in the pathogenesis of gastric carcinoma: Interactions
between genetic alterations, cellular phenotype and cancer
histotype. Hepatogastroenterology. 48:1523–1530. 2001.PubMed/NCBI
|
|
6
|
El-Rifai W and Powell SM: Molecular
biology of gastric cancer. Semin Radiat Oncol. 12:128–140. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang L, Wei D, Huang S, Peng Z, Le X, Wu
TT, Yao J, Ajani J and Xie K: Transcription factor Sp1 expression
is a significant predictor of survival in human gastric cancer.
Clin Cancer Res. 9:6371–6380. 2003.PubMed/NCBI
|
|
8
|
Kurayoshi M, Oue N, Yamamoto H, Kishida M,
Inoue A, Asahara T, Yasui W and Kikuchi A: Expression of Wnt-5a is
correlated with aggressiveness of gastric cancer by stimulating
cell migration and invasion. Cancer Res. 66:10439–10448. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei D, Gong W, Kanai M, Schlunk C, Wang L,
Yao JC, Wu TT, Huang S and Xie K: Drastic down-regulation of
Krüppel-like factor 4 expression is critical in human gastric
cancer development and progression. Cancer Res. 65:2746–2754. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu YL, Chiang YH, Liu GY and Hung HC:
Functional role of dimerization of human peptidylarginine deiminase
4 (PAD4). PLoS One. 6:e213142011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang X and Han J: Expression of
peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol
Carcinog. 45:183–196. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang X, Han J, Pang L, Zhao Y, Yang Y and
Shen Z: Increased PADI4 expression in blood and tissues of patients
with malignant tumors. BMC Cancer. 9:402009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Chang X, Yuan G, Zhao Y and Wang
P: Expression of peptidylarginine deiminase type 4 in ovarian
tumors. Int J Biol Sciences. 6:454–464. 2010. View Article : Google Scholar
|
|
14
|
Ordóñez A, Yélamos J, Pedersen S, Miñano
A, Conesa-Zamora P, Kristensen SR, Stender MT, Thorlacius-Ussing O,
Martínez-Martínez I, Vicente V and Corral J: Increased levels of
citrullinated antithrombin in plasma of patients with rheumatoid
arthritis and colorectal adenocarcinoma determined by a newly
developed ELISA using a specific monoclonal antibody. Thromb
Haemost. 104:1143–1149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simms D, Cizdziel PE and Chomczynski P:
TRIzol: A new reagent for optimal single-step isolation of RNA.
Focus. 15:532–535. 1993.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tiffen JC, Bailey CG, Ng C, Rasko JE and
Holst J: Luciferase expression and bioluminescence does not affect
tumor cell growth in vitro or in vivo. Mol Cancer. 9:2992010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haase SB and Reed SI: Improved flow
cytometric analysis of the budding yeast cell cycle. Cell Cycle.
1:132–136. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Katz M, Amit I, Citri A, Shay T, Carvalho
S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, et al:
A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell
migration. Nat Cell Biol. 9:961–969. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Wysocka J, Sayegh J, Lee YH,
Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y,
et al: Human PAD4 regulates histone arginine methylation levels via
demethylimination. Science. 306:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klose RJ and Zhang Y: Regulation of
histone methylation by demethylimination and demethylation. Nat Rev
Mol Cell Biol. 8:307–318. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cuthbert GL, Daujat S, Snowden AW,
Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD,
Tempst P, Bannister AJ and Kouzarides T: Histone deimination
antagonizes arginine methylation. Cell. 118:545–553. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzuki A, Yamada R, Chang X, Tokuhiro S,
Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono
M, et al: Functional haplotypes of PADI4, encoding citrullinating
enzyme peptidylarginine deiminase 4, are associated with rheumatoid
arthritis. Nat Genet. 34:395–402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang X, Zhao Y, Sun S, Zhang Y and Zhu Y:
The expression of PADI4 in synovium of rheumatoid arthritis.
Rheumatol Int. 29:1411–1416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iwamoto T, Ikari K, Nakamura T, Kuwahara
M, Toyama Y, Tomatsu T, Momohara S and Kamatani N: Association
between PADI4 and rheumatoid arthritis: A meta-analysis.
Rheumatology (Oxford). 45:804–807. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv Y, Xia Y, Wang Y and Cai C: Expression
of PADI4 in hepatocellular carcinoma. Chinese-German J Clin Oncol.
8:453–455. 2009. View Article : Google Scholar
|
|
27
|
Ulivi P, Mercatali L, Casoni GL, Scarpi E,
Bucchi L, Silvestrini R, Sanna S, Monteverde M, Amadori D, Poletti
V and Zoli W: Multiple marker detection in peripheral blood for
NSCLC diagnosis. PLoS One. 8:e574012013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
Outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.PubMed/NCBI
|
|
30
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Turpeenniemi-Hujanen T: Gelatinases (MMP-2
and −9) and their natural inhibitors as prognostic indicators in
solid cancers. Biochimie. 87:287–297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wong TS, Kwong DL, Sham JS, Wei WI, Kwong
YL and Yuen AP: Clinicopathologic significance of plasma matrix
metalloproteinase-2 and −9 levels in patients with undifferentiated
nasopharyngeal carcinoma. Eur J Surg Oncol. 30:560–564. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsumura S, Oue N, Nakayama H, Kitadai Y,
Yoshida K, Yamaguchi Y, Imai K, Nakachi K, Matsusaki K, Chayama K
and Yasui W: A single nucleotide polymorphism in the MMP-9 promoter
affects tumor progression and invasive phenotype of gastric cancer.
J Cancer Res Clin Oncol. 131:19–25. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu GY, Liao YF, Chang WH, Liu CC, Hsieh
MC, Hsu PC, Tsay GJ and Hung HC: Overexpression of peptidylarginine
deiminase IV features in apoptosis of haematopoietic cells.
Apoptosis. 11:183–196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li P, Yao H, Zhang Z, Li M, Luo Y,
Thompson PR, Gilmour DS and Wang Y: Regulation of p53 target gene
expression by peptidylarginine deiminase 4. Mol Cell Biol.
28:4745–4758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao H, Li P, Venters BJ, Zheng S, Thompson
PR, Pugh BF and Wang Y: Histone Arg modifications and p53 regulate
the expression of OKL38, a mediator of apoptosis. J Biol Chem.
283:20060–20068. 2008. View Article : Google Scholar : PubMed/NCBI
|