Introduction
Vascular smooth muscle cells (VSMCs) comprise the
medial layer of blood vessels, control vessel tone and blood flow,
and thereby serve a fundamental role in blood pressure regulation
and substance exchange (1,2). Under pathological conditions, VSMCs
typically exhibit increased proliferative, migratory and
extracellular matrix-synthesizing capacities, indicating that they
convert to the synthetic phenotype (3). Autophagy is a cellular catabolic
process responsible for the destruction of long-lived proteins and
organelles via a lysosome-dependent pathway that maintains cellular
homeostasis. Deregulated autophagy has been implicated in the
pathogenesis of various diseases, including vascular disorders
(4). A number of in vitro
studies have demonstrated that various pro-atherogenic stimuli are
able to induce autophagy in vascular cells (5,6). A
recent study indicated that there may be a correlation between
autophagy and hypoxia in the development and progression of
vasculopathy (4). In pulmonary
vascular cells, autophagy activation inhibits proliferation when
cells are exposed to hypoxic conditions. Hypoxia has been
demonstrated to activate autophagy via regulation of adenosine
monophosphate-activated protein kinase (AMPK) in human pulmonary
smooth muscle cells. Furthermore, suppressing AMPK expression
prevents hypoxia-mediated autophagy and the induction of cell death
(7). In human umbilical vein
endothelial cells (HUVECs), autophagy is induced when cells are
subjected to hypoxic conditions, which is enhanced by
hypoxia-inducible factor 1 (HIF-1) gene overexpression and
inhibited by HIF-1 loss-of-function (8). However, the extent to which molecular
mechanisms of hypoxia contribute to the induction of autophagy in
VSMCs remains to be determined.
MicroRNA (miR) are small non-coding RNA that serve
as important post-transcriptional gene regulators, with a primary
function of controlling cell proliferation and differentiation of
various cell types. A number of previous studies have demonstrated
that pathogenic changes in tissues, including cardiac hypertrophy,
heart failure, cardiac fibrosis and vascular atherosclerosis, may
be associated with miR (9,10). A recent study has indicated that
miR-137 inhibits hypoxia-induced mitophagy by reducing expression
of the mitophagy receptor, thereby leading to inadequate
interaction between the receptor and light chain (LC) 3 (11), further indicating that miR has a role
in modulating autophagy. Furthermore, it has been demonstrated that
miR-20a-5p mediates hypoxia-induced autophagy by targeting the
autophagy related 16-like 1 gene in ischemic kidney injury
(12), and in cardiomyocytes, the
inhibition of miR-497 is able to ameliorate anoxia/reoxygenation
injury by suppressing cell apoptosis and enhancing autophagy
(13). miR-17-5p is a member of the
miR-17–92 cluster, which is located on human chromosome 13q31
(14). A previous study revealed the
widespread overexpression of miR-17-5p in diverse tumor tissues,
suggesting that miR-17-5p exhibits oncogenic activity (15). Notably, miR-17-5p promotes
oxidative-stress-induced cardiomyocyte apoptosis in animal models
of ischemia/reperfusion-induced cardiac injury and cellular models
of cardiomyocyte injury (16).
However, the roles of miR-17-5p in hypoxia-induced autophagy and
apoptosis in VSMCs have not been investigated until recently.
The present study aimed to demonstrate that
miR-17-5p is a regulator of signal transducer and activator of
transcription 3 (STAT3) by using miRanda, TargetScan and picTar
databases, and to elucidate if miR-17-5p is able to mediate
hypoxia-induced autophagy and inhibit apoptosis by targeting STAT3
in VSMCs.
Materials and methods
Cell culture
VSMCs were obtained from the Cell Resource Center,
Shanghai Institutes for Biological Sciences (Shanghai, China) and
maintained in RPMI-1640 supplemented with 10% fetal bovine serum
(FBS; both Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) at 37°C in a humidified incubator (Thermo Fisher Scientific,
Inc.), in an atmosphere containing 5% CO2. Ethyl
3,4-dihydroxybenzoate (EDHB) was purchased from Sigma-Aldrich;
Merck Millipore (Darmstadt, Germany) and dissolved in ethanol.
VSMCs were exposed to EDHB (50 µg/ml) for 24 h.
Caspase-3 activity and cell apoptosis
assay
VSMC lysates were prepared and measured using a
caspase-3 ELISA Kit (no. KGA203, Nanjing KeyGEN BioTECH, Co., Ltd.,
Nanjing China). In brief, 50 µl supernatant was mixed with 2X
reaction buffer (50 µl) and dithiothreitol (0.5 µl).
Immunocomplexes were incubated with 5 µl peptide substrate in assay
buffer for 2 h at 37°C. Release of p-nitroaniline was measured at
405 nm using an ELISA reader (SpectraMax M5; Molecular Devices,
LLC., Sunnyvale, CA, USA) according to the manufacturer's
instructions.
Quantitative assessment of apoptotic cells was
performed using the terminal deoxynucleotidyl transferase dUTP nick
end labeling (TUNEL) method, examining DNA-strand breaks during
apoptosis with the ApoAlert DNA Fragmentation Assay kit (BD
Biosciences, Franklin Lakes, NJ, USA). Briefly, cells were
incubated at 37°C in hypoxic conditions for 48 h. Cells were
trypsinized, fixed with 4% paraformaldehyde at room temperature for
24 h and permeabilized with 0.1% Triton-X-100 in 0.1% sodium
citrate. Cells were washed and incubated with the reaction mixture
for 60 min at 37°C, and immediately analyzed using FACScan and the
Cellquest program ver. 5.1 (BD Biosciences).
Overexpression and small interfering
(si) RNA
Lentiviral vectors containing miR-17-5p were
constructed to transfect VSMCs. Briefly, VSMCs were cultured in
McCoy's 5α medium containing 10% FBS (MP, Biomedicals, Santa Ana,
CA, USA) and when the exponential growth phase was reached,
1.0×105 cells/well were seeded in 96-well plates. A
total of 300 µl complete culture medium, containing recombinant
lentiviruses, control lentiviruses or McCoy's 5α medium (all of
which contained 6 µg/ml polybrene; Sigma-Aldrich; Merck Millipore)
was added to the plates when the cells reached 50–60% confluence.
The virus-containing medium was replaced with fresh complete medium
two days later.
The siRNAs or antagomirs for miR-17-5p, were
obtained from GE Healthcare Dharmacon, Inc. (Pittsburgh, PA, USA),
and were designed with the following sequences: miR-17-5p,
5′-CUGAGGUCCAGGACACACA-3′; scramble, 5′-AGAGAUGACUCACUGUCAC-3′.
VSMCs were transfected with siRNA oligonucleotides using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
HIF-1α-siRNA transfection
The HIF-1α-siRNA oligonucleotide fragment was
designed and synthesized according to the human HIF-1α gene
sequence (GenBank no. NM001530). The sequence was as follows:
Forward,
5′-GATCCCGAGGAAGAACTATGAACATAATTCAAGAGATTATGTTCATAGTTCTTCCTCTTTTTGGAT-3′
and reverse,
5′-AGCTATCCAAAAAGAGGAAGAACTATGAACATAATCTCTTGAATTATGTTCATAGTTCTTCCTCGG-3′.
Scramble and HIF-1α-siRNA oligonucleotide were cloned into the
pSIREN-RetroQ plasmid (Addgene, Inc., Cambridge, MA, USA) for
retrovirus production. After 48 h, infected cells were selected
with puromycin (2 mg/ml) and the clones were selected and cultured
for further experiment.
Luciferase reporter gene activity
assay
The three prime untranslated region (3′UTR) of the
STAT3 gene containing the predicated target sites for miR-17-5p was
obtained by polymerase chain reaction (PCR) amplification. The
fragment was inserted into the multiple cloning sites of the
pMIR-REPORT luciferase miR expression reporter vector (Ambion;
Thermo Fisher Scientific, Inc.). Cells from the human embryonic
kidney cell line HEK-293 (Cell Resource Center, Shanghai Institutes
for Biological Sciences) were co-transfected with 0.1 µg luciferase
reporters containing STAT3 3′UTR and microR-17-5p mimics using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell lysates were harvested 48 h post-transfection and luciferase
activity was measured with a dual luciferase reporter assay kit
(no. RG028. Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA extraction was performed using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Synthesis of cDNA was performed by RT
reactions with 2 µg total RNA using Moloney murine leukemia virus
reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.)
with oligo dT (15) primers
(Fermentas; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. miR-17-5p level was measured using qPCR
with the mirVana RT-qPCR miR detection kit (Ambion; Thermo Fisher
Scientific, Inc.) in conjunction with SYBR Green (Invitrogen;
Thermo Fisher Scientific, Inc.). Following the circle reaction, the
threshold cycle (Cq) was determined and the relative miR-17-5p
level was calculated based on the Cq values and normalized to U6
level in each sample. PCR was performed with the following primers:
miR-17-5p, forward 5′-TCTAGATCCCGAGGACTG-3′ and reverse,
5′-ATCGTGACCTGAACC-3′; U6, forward 5′-CTCGCTTTGGCAGCACA-3′ and
reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Autophagy detection in VSMCs
Autophagy in VSMCs was detected via western blotting
and fluorescence microscopy, and cultured VSMCs were prepared for
observation under a transmission electron microscope (TEM) to
investigate the formation of autophagosomes. Cells were washed with
phosphate-buffered saline, fixed with 4% formaldehyde/1%
glutaraldehyde at room temperature overnight, and then rinsed three
times with 0.1 M sodium cacodylate/0.2 M sucrose buffer. Following
fixation, cells were incubated in 1% osmium tetroxide for 1 h,
dehydrated in a series of ethanol washes, and then embedded in
epoxy resin. When the epoxy resin had polymerized at 65°C for 24 h,
sections 100-nm thick were then cut onto a carbon-coated copper
grid. Cells were stained with uranyl acetate and lead citrate (SPI
Supplies, West Chester, PA, USA) and examined under a TEM (H-800;
Hitachi, Ltd., Tokyo, Japan).
Western blotting
VSMCs were homogenized and extracted in NP-40 buffer
(Thermo Fisher Scientific, Inc.), followed by 5–10 min boiling and
centrifugation at 7,500 × g, for 15 min at 4°C to obtain the
supernatant. Protein samples were quantified using the
bicinchoninic Acid kit for Protein Determination, (no. BCA1-1KT;
Sigma-Aldrich; Merck Millipore).
Samples containing 30 µg protein were separated by
10% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Following blocking for 2 h
at room temperature with 5% (w/v) non-fat dry milk in tris-buffered
saline and 0.1% (w/v) Tween 20 (TBST), membranes were incubated
with primary antibodies for LC3 (sc-292,354, dilution, 1:1,000),
P62 (sc-48389, dilution, 1:1,000), β-actin (sc-81178, dilution,
1:2,000), Bax (sc-6236, dilution, 1:1,000), p-caspase3 (sc-22171-R,
dilution, 1,000), caspase3 (sc-7272, dilution, 1:1,000), STAT2
(sc-483, dilution, 1:1,000) at 4°C overnight. All primary
antibodies were purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). The membranes were washed three times with TBST
and incubated with secondary antibodies donkey anti-mouse
immunoglobulin (Ig) G (sc-2096, dilution, 1:10,000) and goat
anti-rabbit IgG (sc-2004, dilution, 1:10,000; both Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature and visualized
with an Amersham ECL Western blotting Detection reagent (GE
Healthcare Life Sciences, Chalfont, UK). Signals were
densitometrically assessed using Quantity One® software
ver. 4.5 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each
experiment was performed four times.
Statistical analysis
The data from these experiments were presented as
the mean ± standard deviation for each group. Statistical analyses
were performed using PRISM version 5.0 (GraphPad Software, Inc., La
Jolla, CA, USA). Inter-group differences were analyzed via one-way
analysis of variance. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-17-5p expression in response to
hypoxia and inhibitors
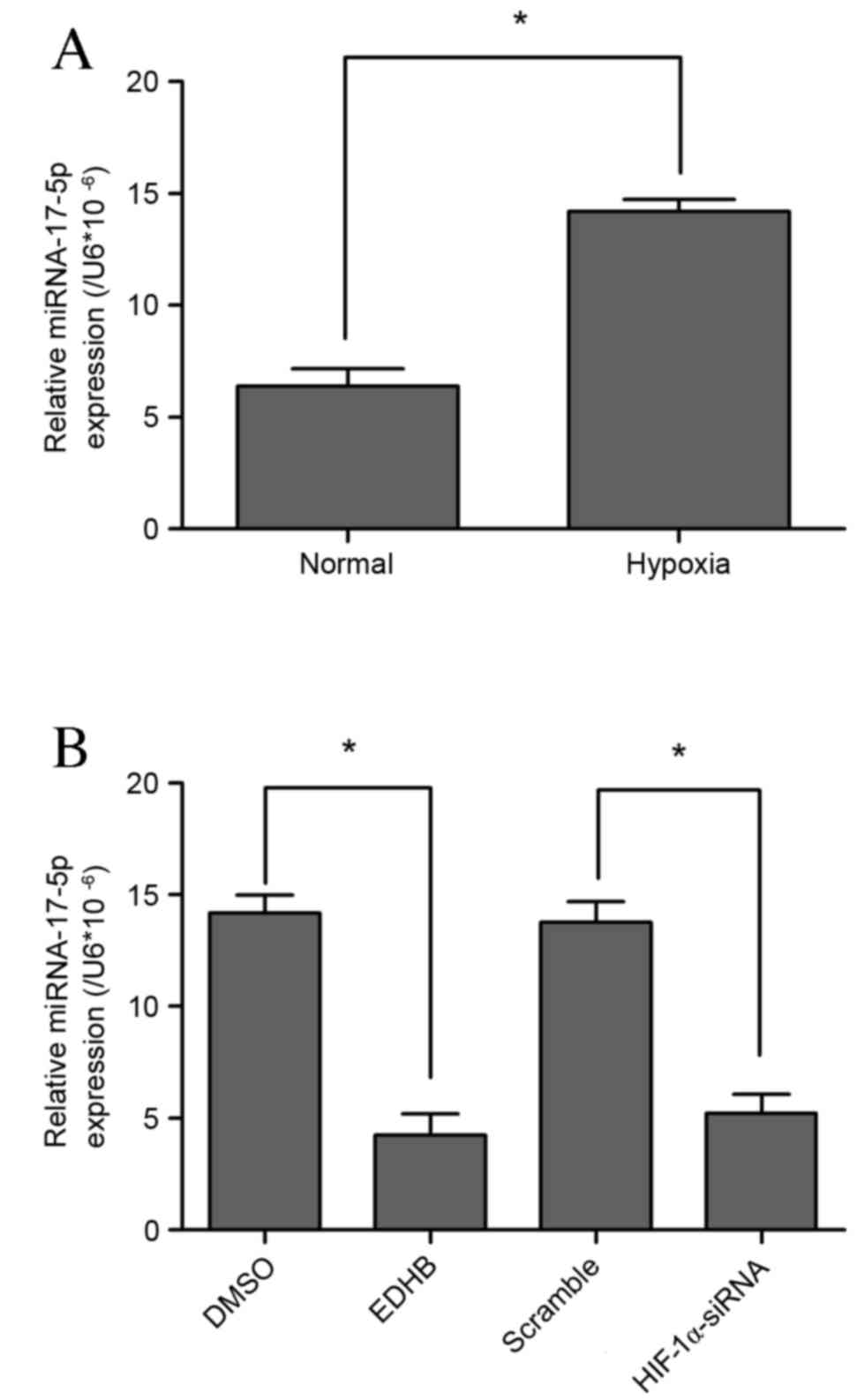
Significant upregulation of miR-17-5p expression was
observed in VSMCs subjected to hypoxic conditions (P<0.05;
Fig. 1A). HIF-1 is stabilized under
hypoxic conditions as a cellular response to low oxygen
concentration. Furthermore, EDHB has been shown to have a
cytoprotective effect against oxidative stress in various types of
cells, including VSMCs (17). In the
present study, lower miR-17-5p levels were observed in EDHB-treated
and HIF-1α loss-of-function cells (Fig.
1B). These results indicate that miR-17-5p may be a novel
hypoxia-responsive miR that has not been reported in previous miR
profiling data.
Hypoxia induces autophagy and inhibits
apoptosis in VSMCs
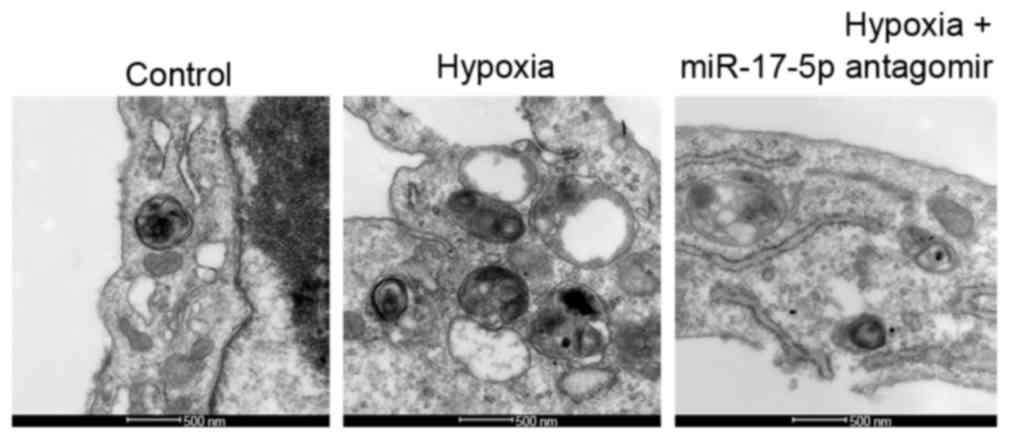
The ultrastructure of VSMCs was observed under a TEM
to assess levels of autophagy in response to hypoxia and miR-17-5p
loss-of-function. An increase in typical autophagic vacuoles
containing extensively degraded organelles, including mitochondria
and endoplasmic reticulum, was observed in the cytoplasm of VSMCs
following treatment with hypoxia as compared with the control group
(Fig. 2). However, this effect was
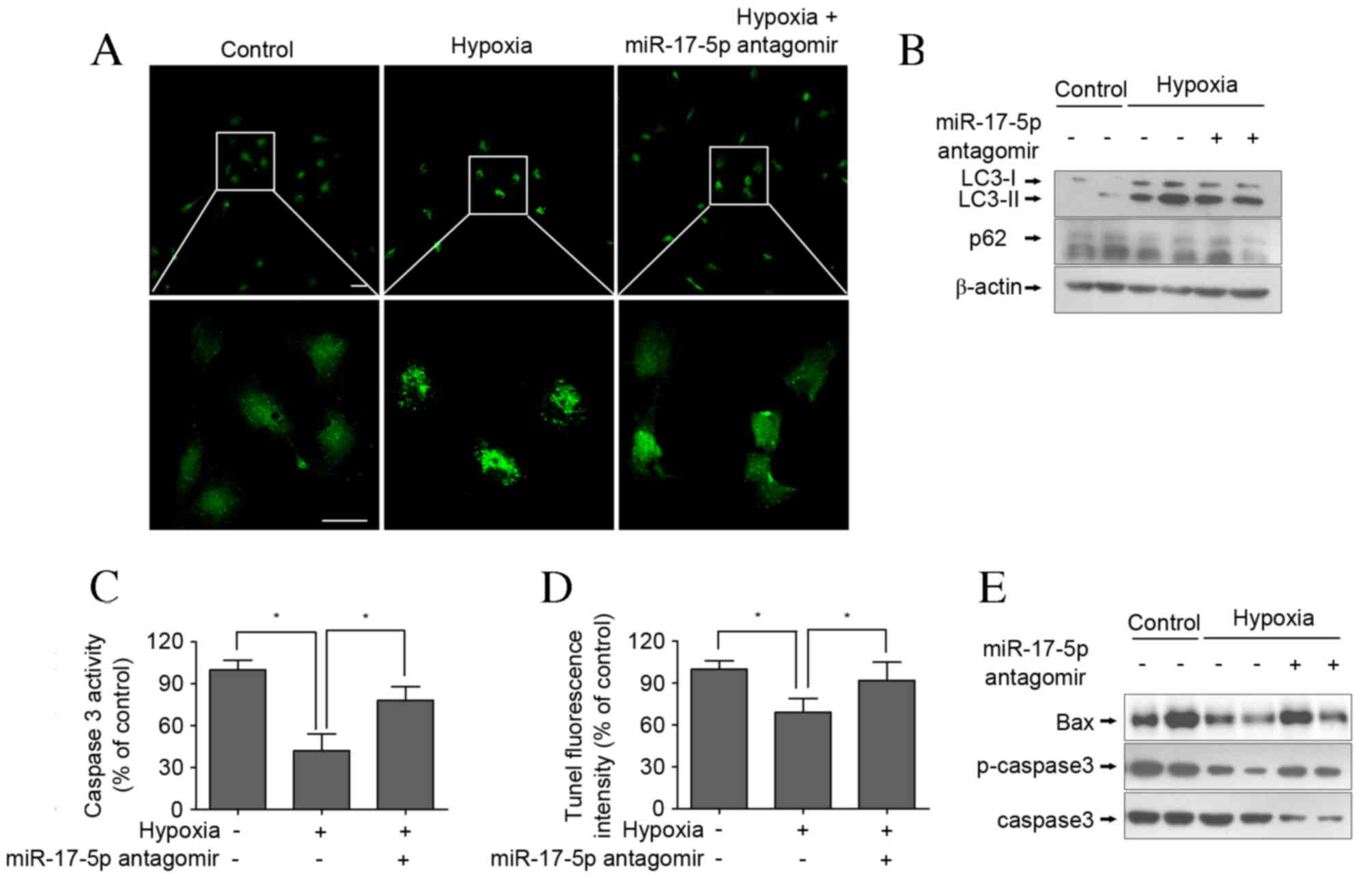
reversed by miR-17-5p antagomir in hypoxic conditions (Fig. 2). To assess autophagosome formation,
LC3-II expression was measured via immunofluorescent staining and
western blotting was used to measure the LC3-II/LC3-I ratio and p62
levels. The results demonstrated that the LC3-II intensity of green
fluorescence in response to hypoxia was greater than that of the
control group following incubation for 24 h in RPMI-1640 (Fig. 3A). Consistent with the fluorescence
imaging results, the LC3-II/LC3-I ratio increased in response to
hypoxia (Fig. 3B) and the expression
of p62 was markedly decreased following exposure of cells to
hypoxic conditions. However, the results of LC3-II degeneration and
p62 accumulation demonstrated that miR-17-5p loss-of-function
inhibited hypoxia-induced autophagy (Fig. 3A and B). Furthermore, it was
investigated whether hypoxia induced apoptosis in VSMCs via an
apoptotic mechanism using a caspase-3 activity assay and TUNEL
staining following the exposure of VSMCs for 24 h. The results
indicate that caspase-3 activity in the hypoxia-treated group was
significantly lower than that of the control group (P<0.05;
Fig. 3C), and cell apoptosis was
significantly inhibited in response to hypoxia (P<0.05; Fig. 3D); however, these reductions were
significantly reversed by miR-17-5p antagomir transfection (both
P<0.05). The apoptotic response was further investigated by
measuring the expression of apoptosis-related proteins. Western
blot analysis demonstrated that treatment of VSMCs with hypoxia
markedly decreased pro-apoptotic proteins BAX and p-caspase levels
(Fig. 3E). These results suggest
that hypoxia induced protective autophagy in VSMCs via inhibition
of cell apoptosis as a response to an anaerobic environment, which
is consistent with the literature (18).
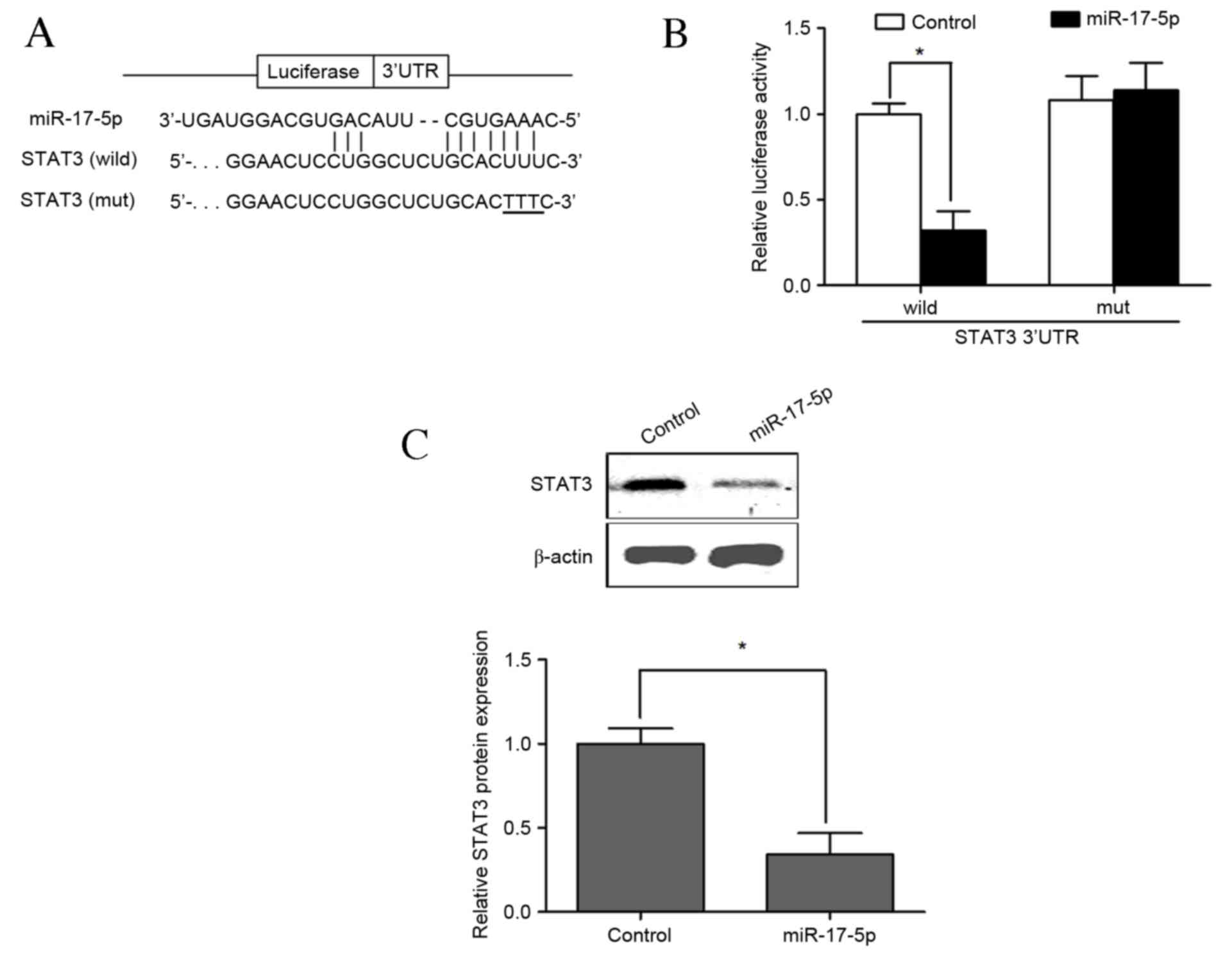
miR-17-5p directly targets the 3′-UTR
of STAT3
To determine whether miR-17-5p regulated STAT3
through the predicted binding sites in its 3′-UTR (Fig. 4A), a luciferase construct was
designed by incorporating wild-type or mutant 3′-UTR of STAT3,
which expressed luciferase unless repressed by the incorporated
3′-UTR. No significant difference was observed between the control
group and the group that underwent cotransfection of VSMCs with the
pMIR-REPORT construct containing mutant STAT3 3′-UTR and
PLemiR-17-5p. Cotransfection with the luciferase construct
containing wild-type STAT3 3′-UTR and PLemiR-17-5p resulted in
significantly lower luciferase activity than the control group,
leading to a ~70% decline in luciferase activity compared with
control group (P<0.05; Fig. 4B).
Western blotting results indicated that the expression of STAT3 was
significantly decreased following cotransfection with the
luciferase construct containing wild-type STAT3 3′-UTR and
overexpressing miR-17-5p compared with that of the wild-type
control group (P<0.05; Fig. 4C).
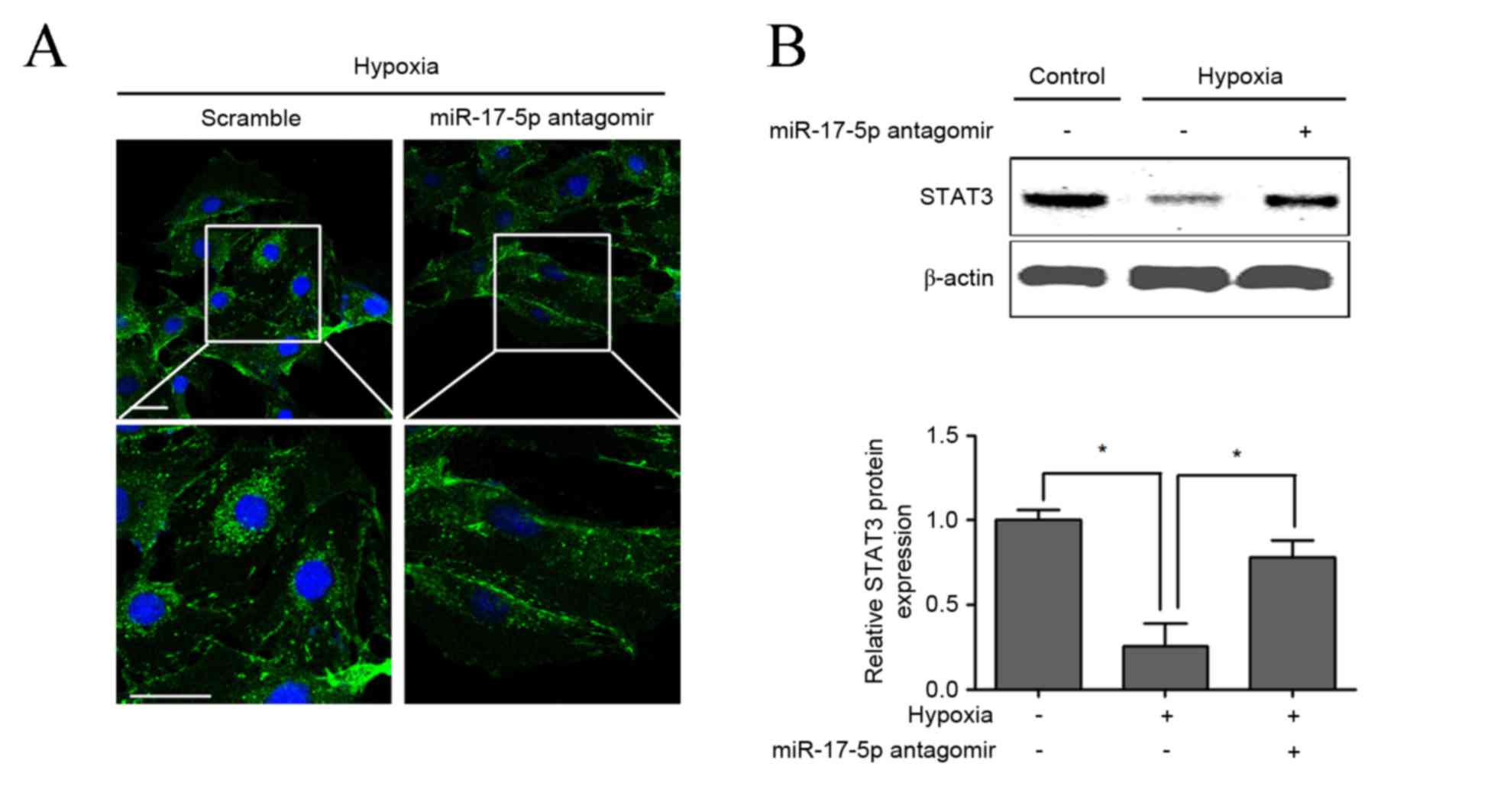
Furthermore, the combination of miR-17-5p loss-of-function and
hypoxia was demonstrated to significantly upregulate the protein
expression of STAT3 compared with hypoxia single treatment in VSMCs
(P<0.05), which was confirmed by immunofluorescent staining
(Fig. 5A) and western blotting
(Fig. 5B).
Discussion
The present study identified miR-17-5p as a novel
hypoxia-responsive miR of VSMC autophagy and apoptosis via
targeting STAT3. MiR-17-5p was significantly upregulated in
response to hypoxia, which also significantly increased autophagic
vacuoles and decreased apoptosis. However, miR-17-5p
loss-of-function was able to reverse hypoxia-induced increased
autophagic vacuoles and decreased apoptosis, indicating that
miR-17-5p may be associated with hypoxia-induced protective
autophagy and anti-apoptosis in VSMCs.
Autophagy has dual roles in cardiovascular disease:
Physiological autophagy serves as a protective mechanism to
maintain normal cardiovascular function, whereas impaired autophagy
contributes to the development and progression of various diseases
(19). A previous study indicated
that in cardiac HL-1 cells, ischemia/reperfusion-induced apoptosis
is decreased when autophagy is increased, suggesting that it serves
a cardioprotective role in response to cardiac injury (20). Conversely, pro-apoptosis may
synergistically increase autophagy in oxidated low-density
lipoprotein-treated VSMCs via the suppression of miR hsa-let-7g
(5). In HUVECs, HIF-1 is able to
reduce cell viability by inducing autophagy (8). The present study demonstrated that
increasing autophagy may inhibit apoptosis in hypoxia-treated
VSMCs, suggesting that autophagy may serve as a protective
mechanism for hypoxia-treated VSMCs via the inhibition of cell
apoptosis.
miR-17-5p is a member of the microR-17–92 cluster
and is associated with the regulation of tumor proliferation and
progression (21,22). Notably, miR-17-5p is associated with
ischemia/reperfusion and cardiomyocyte injury under oxidative
stress in mice, and overexpression of miR-17-5p aggravates
cardiomyocyte injury leading to reduced cell viability and enhanced
H2O2-induced apoptotic cell death, whereas
inhibition of miR-17-5p by its anti-miR oligonucleotide, AMO-17-5p,
abrogates the deleterious changes (16). The present study demonstrated that
miR-17-5p is upregulated in VSMCs when cells are exposed to hypoxia
and, furthermore, that miR-17-5p antagomir was able to restore the
LC3-II/LC3-I ratio, which increases in response to hypoxia in
VSMCs. The conversion of LC3-I into LC3-II is an essential step in
autophagosome formation and the abundance of LC3-II is correlated
with the number of autophagosomes (5,18). These
results indicated that LC3-II degenerates in response to miR-17-5p
loss-of-function inhibited hypoxia-induced autophagy.
It is known that STAT3 is required to regulate cell
proliferation, differentiation, and apoptosis, and activation of
STAT3 by phosphorylation increases cell survival in response to
various cytokines and stress stimuli (23). In VSMCs, STAT3 is associated with
cell senescence and calcification, and STAT3 expression is
significantly decreased in response to miR-135a overexpression
(24). Recent in vitro and
in vivo research has demonstrated that miR-17-5p
overexpression promotes cardiomyocyte apoptosis, which is induced
by oxidative stress via the targeting of STAT3 (16). In the present study, hypoxia-induced
miR-17-5p upregulation was able to inhibit STAT3 expression in
vitro; therefore STAT3, as a target for miR-17-5p, may be
associated with hypoxia-induced protective autophagy.
In conclusion, the present study demonstrated that
miR-17-5p is associated with hypoxia-induced protective autophagy
and apoptosis and that the underlying mechanism was mediated, at
least partially, by targeting STAT3.
Acknowledgements
This study was supported by the National Natural
Science Foundation for the Youth of China (grant no.81500198)
References
|
1
|
Salabei JK and Hill BG: Autophagic
regulation of smooth muscle cell biology. Redox biology. 4:97–103.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Meyer GR, Grootaert MO, Michiels CF,
Kurdi A, Schrijvers DM and Martinet W: Autophagy in vascular
disease. Circ Res. 116:468–479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boya P, Reggiori F and Codogno P: Emerging
regulation and functions of autophagy. Nat Cell Biol. 15:713–720.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vindis C: Autophagy: An emerging
therapeutic target in vascular diseases. Br J Pharmacol.
172:2167–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding Z, Wang X, Schnackenberg L, Khaidakov
M, Liu S, Singla S, Dai Y and Mehta JL: Regulation of autophagy and
apoptosis in response to ox-LDL in vascular smooth muscle cells,
and the modulatory effects of the microRNA hsa-let-7g. Int J
Cardiol. 168:1378–1385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng N, Meng N, Wang S, Zhao F, Zhao J, Su
L, Zhang S, Zhang Y, Zhao B and Miao J: An activator of mTOR
inhibits oxLDL-induced autophagy and apoptosis in vascular
endothelial cells and restricts atherosclerosis in apolipoprotein
E-/− mice. Sci Rep. 4:55192014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ibe JC, Zhou Q, Chen T, Tang H, Yuan JX,
Raj JU and Zhou G: Adenosine monophosphate-activated protein kinase
is required for pulmonary artery smooth muscle cell survival and
the development of hypoxic pulmonary hypertension. Am J Respir Cell
Mol Biol. 49:609–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu J, Lei Z and Yu J: Hypoxia induces
autophagy in human vascular endothelial cells in a
hypoxia-inducible factor 1dependent manner. Mol Med Rep.
11:2677–2682. 2015.PubMed/NCBI
|
|
9
|
Pan ZW, Lu YJ and Yang BF: MicroRNAs: A
novel class of potential therapeutic targets for cardiovascular
diseases. Acta Pharmacol Sin. 31:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Urbich C, Kuehbacher A and Dimmeler S:
Role of microRNAs in vascular diseases, inflammation, and
angiogenesis. Cardiovasc Res. 79:581–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Zhang X, Zhuang H, Chen HG, Chen Y,
Tian W, Wu W, Li Y, Wang S, Zhang L, et al: MicroRNA-137 is a novel
hypoxia-responsive microRNA that inhibits mitophagy via regulation
of two mitophagy receptors FUNDC1 and NIX. J Biol Chem.
289:10691–10701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang IK, Sun KT, Tsai TH, Chen CW, Chang
SS, Yu TM, Yen TH, Lin FY, Huang CC and Li CY: MiR-20a-5p mediates
hypoxia-induced autophagy by targeting ATG16L1 in ischemic kidney
injury. Life Sci. 136:133–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Zeng Z, Li Q, Xu Q, Xie J, Hao H,
Luo G, Liao W, Bin J, Huang X and Liao Y: Inhibition of
microRNA-497 ameliorates anoxia/reoxygenation injury in
cardiomyocytes by suppressing cell apoptosis and enhancing
autophagy. Oncotarget. 6:18829–18844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu J, Ohuchida K, Mizumoto K, Fujita H,
Nakata K and Tanaka M: MicroRNA miR-17-5p is overexpressed in
pancreatic cancer, associated with a poor prognosis, and involved
in cancer cell proliferation and invasion. Cancer Biol Ther.
10:748–757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Du W, Pan Z, Chen X, Wang L, Zhang Y, Li
S, Liang H, Xu C, Zhang Y, Wu Y, et al: By targeting Stat3
microRNA-17-5p promotes cardiomyocyte apoptosis in response to
ischemia followed by reperfusion. Cell Physiol Biochem. 34:955–965.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nimker C, Kaur G, Revo A, Chaudhary P and
Bansal A: Ethyl 3,4-dihydroxy benzoate, a unique preconditioning
agent for alleviating hypoxia-mediated oxidative damage in L6
myoblasts cells. J Physiol Sci. 65:77–87. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee J, Giordano S and Zhang J: Autophagy,
mitochondria and oxidative stress: Cross-talk and redox signalling.
Biochem J. 441:523–540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mei Y, Thompson MD, Cohen RA and Tong X:
Autophagy and oxidative stress in cardiovascular diseases. Biochim
Biophys Acta. 1852:243–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hamacher-Brady A, Brady NR and Gottlieb
RA: Enhancing macroautophagy protects against ischemia/reperfusion
injury in cardiac myocytes. J Biol Chem. 281:29776–29787. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Spaccarotella E, Pellegrino E, Ferracin M,
Ferreri C, Cuccuru G, Liu C, Iqbal J, Cantarella D, Taulli R,
Provero P, et al: STAT3-mediated activation of microRNA cluster
17~92 promotes proliferation and survival of ALK-positive
anaplastic large cell lymphoma. Haematologica. 99:116–124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Novotny GW, Sonne SB, Nielsen JE, Jonstrup
SP, Hansen MA, Skakkebaek NE, Rajpert-De Meyts E, Kjems J and
Leffers H: Translational repression of E2F1 mRNA in carcinoma in
situ and normal testis correlates with expression of the miR-17–92
cluster. Cell Death Differ. 14:879–882. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao XH, Wang N, Zhao DW, Zheng DL, Zheng
L, Xing WJ, Ma WJ, Bao LY, Dong J and Zhang TC: STAT3 Protein
Regulates Vascular Smooth Muscle Cell Phenotypic Switch by
Interaction with Myocardin. J Biol Chem. 290:19641–19652. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin L, He Y, Xi BL, Zheng HC, Chen Q, Li
J, Hu Y, Ye MH, Chen P and Qu Y: miR-135a suppresses calcification
in senescent VSMCs by regulating KLF4/STAT3 pathway. Curr Vasc
Pharmacol. 14:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|