Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a
common pathogen in hospital-acquired infections (1). Due to increasing multidrug resistance,
P. aeruginosa infection is an increasingly common cause of
mortality and morbidity (2). The
mechanisms of antibiotic resistance in P. aeruginosa include
the expression of multiple antibiotic modifying enzymes, antibiotic
efflux pumps and acquisition of chromosomally or plasmid encoded
antibiotic resistance genes. Additionally, chromosomal mutations
and lower membrane permeability for the antibiotics also contribute
to antibiotic resistance (3). P.
aeruginosa is a biofilm-forming pathogen and is difficult to
eradicate due to its high antibiotic resistance and the ability of
the biofilm to evade the immune system (4–7). Quorum
sensing (QS) is a system of stimuli and response correlated to
population density. P. aeruginosa uses the QS system to
coordinate gene expression according to the density of its local
population. Thus, it can coordinate certain behaviors such as
biofilm formation, virulence and antibiotic resistance. QS
inhibitors (QSIs) are the most well reported alternative
therapeutics that can be used to overcome the problem of increasing
antibiotic resistance in P. aeruginosa. QSIs target the
virulence of the organism and therefore are also termed
antipathogenic drugs. The virulence of P. aeruginosa depends
on its cell-to-cell communication system, or QS system that uses
diffusible signaling molecules that accumulate with increasing cell
density and allows P. aeruginosa to trigger coordinated
responses and achieve outcomes that would otherwise remain
impossible to achieve by individual bacterium (8). Previous studies have demonstrated that
traditional treatments for bacteria exert some effect on biofilm
infections (9,10). Therefore, the effects of constituents
from marine organisms, traditional Chinese herbs and plants
(11–13) on biofilm infections are being
assessed.
Flavonoids are plant polyphenols present in
vegetables, fruits and beverages of plant origin and are well known
for their antipyretic, analgesic and anti-inflammatory
physiological properties (14–18).
Hyperoside is a type of modified flavonoid. It has been
demonstrated that hyperoside has weak antibacterial activity
against gram-positive bacteria and no antibacterial activity
against gram-negative bacteria (19,20).
Furthermore, it has been identified that hyperoside exhibits an
inhibitory effect on P. aeruginosa PAO1 (PAO1) biofilms
(21). While the incidence of
infections caused by antibiotic resistant strains has increased,
the discovery of novel classes of antibiotics has slowed down which
made it imperative to search for alternative treatment strategies.
Therefore, the current study examined the biofilm formation,
adhesion and motility of PAO1 in the presence of hyperoside. And
the hyperoside may become a more effective method to treat the
infection of P. aeruginosa.
Materials and methods
Bacterial strains and culture
conditions
P. aeruginosa PAO1 was acquired from Bioplus
Biotech Co., Ltd. (Shanghai, China) and cultured in Lubria-Bertani
(LB) medium (BD Diagnostics, Sparks Glencoe, MD, USA) at 37°C for
24 h in all experiments. The hyperoside was purchased from Dalian
Meilun Biotechnology Co., Ltd. (Dalian, China).
Dose effect of hyperoside on biofilm
formation
PAO1 was activated in LB medium overnight at 37°C
prior to 1:1,000 (v/v) dilution in a tissue culture microtiter
plate. The final concentrations of hyperoside in the tissue culture
microtiter plate were 8, 16, 32, 64, 128 and 256 µg/ml. Following
24 h incubation at 37°C, the medium was removed and wells were
washed three times with ddH2O. The microtiter plate was
dried prior to the addition of 1% crystal violet (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany) for 15 min at room
temperature. Following staining, the dye was removed and the wells
were washed three times with tap water. The microtiter plate was
dried prior to the addition of 30% glacial acetic acid to
solubilize the dye bound to the biofilm. The absorbance was
measured by xMark Microplate Spectrophotometer (Bio-Rad,
Laboratories, Inc., Hercules, CA, USA) at 590 nm. The optimal
concentration for biofilm inhibition was used in the later
experiments.
Growth assays
Cells were grown in LB medium in the presence or
absence of 16 µg/ml hyperoside. Bacterial culture turbidity was
measured by xMark Microplate Spectrophotometer (Bio-Rad,
Laboratories, Inc) at 600 nm at intervals of 0 h up to 24 h.
Microscopy analysis
Confocal laser scanning microscopy (CLSM) was
performed to analyze the effect of hyperoside on the PAO1 biofilm
at 24 h. Prior to the CLSM experiments, cell cultures were divided
into control and hyperoside (16 µg/ml)-treated groups. Biofilms on
the culture dish were fixed with 2.5% glutaraldehyde at room
temperature for 3 h. Following washing with phosphate-buffered
saline (PBS), 5 µg/ml propidium iodide (Sigma-Aldrich) was added
and the biofilms were incubated for 15 min at 4°C. The plate was
then washed, 50 µg/ml fluorescein isothiocyanate-concanavalin A
(Sigma-Aldrich) was added and the plate was incubated for 30 min at
4°C. The stained biofilm was then observed using CLSM.
Adhesion assays
Adhesion assays were performed as previously
described with minor modifications (22). Following PAO1 activation, the control
and hyperoside groups were cultured in a 96-well microtiter plate
and incubated for 4 h at 37°C. Following incubation, the attached
cells were stained with filtered 1% crystal violet (Sigma-Aldrich)
at room temperature for 15 min. The dye was dissolved in 30%
glacial acetic acid (Sigma-Aldrich), and the absorbance was
measured by xMark Microplate Spectrophotometer at 590 nm.
Motility assays
Twitching motilities were assayed on agar plates
(freshly prepared LB agar plates with 1% Bacto agar were used for
the twitching assay) in the presence or absence of 16 µg/ml
hyperoside (23). An overnight
culture was stabbed with a toothpick to transfer PAO1 through the
agar layer (point-incubation) to the bottom of the Petri dish and
plates were then incubated at 37°C for 48 h. The agar was removed
and attached cells were stained with 1% crystal violet
(Sigma-Aldrich) at room temperature for 15 min. The plates were
washed gently with PBS to remove unattached cells prior to
staining. The diameter of the stained zone was measured by
graduated scale to assess the twitching motility.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect the transcription levels
of lasI, lasR, rhlI and rhlR genes in P. aeruginosa with or
without 16 µg/ml hyperoside. The primers used to amplify these
genes are listed in Table I. Total
RNA was isolated from PAO1 using a FastRNA Pro Blue Kit (MP
Biomedicals, Santa Ana, CA, USA). Cells were grown overnight at
37°C in the presence or absence of hyperoside and harvested by
centrifugation at 16,100 × g for 10 min, and the deposit was
resuspended in TRIzol (Tiangen Biotech Co., Ltd., Beijing, China).
Preparation of total RNA was performed according to the
manufacturer's protocol. The RNA sample was treated with 50 units
of DNase I (Roche Applied Science, Penzberg, Germany) for 2 h at
37°C to remove contaminating DNA. DNaseIwas eliminated by
phenol-chloroform extraction and ethanol precipitation. The pellet
was resuspended in diethyl pyrocarbonate (DEPC)-treated
H2O. Reverse transcription was performed using the First
Strand cDNA Synthesis kit (Toyobo Co., Ltd., Osaka, Japan) and qPCR
using SYBR® Green Real-time PCR Master mix (Toyobo Co., Ltd.)
according to the manufacturer's protocol. The reaction procedure
involved two-step PCR programme: 94°C for 5 min, (94°C for 30 sec,
57°C for 30 sec and 72°C for 30 sec) X40 cycles. The 16S rRNA gene
was selected as the internal control to normalize the data.
Relative expression of gene (RQ) was calculated by
2−∆∆cq and percent reduction was calculated as (1-RQ) X
100 (24). Experiments were repeated
independently three times.
 | Table I.Primer sequences used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer
direction | Sequence
(5′-3′) |
|---|
| lasR | Forward |
CTGTGGATGCTCAAGGACTAC |
|
| Reverse |
ACCGAACTTCCGCCGAAT |
| lasI | Forward |
CGTGCTCAAGTGTTCAAGGA |
|
| Reverse |
GCGTCTGGATGTCGTTCTG |
| rhlR | Forward |
CCGATGCTGATGTCCAACC |
|
| Reverse |
GCTACATCGTCGCCATGAG |
| rhlI | Forward |
GCTACATCGTCGCCATGAG |
|
| Reverse |
TCTCGCCCTTGACCTTCTG |
| 16S | Forward |
ATCTTCGGACCTCACGCTATC |
|
| Reverse |
CCAACTTGCTGAACCACCTAC |
Statistical analysis
The results are expressed as the mean ± standard
deviation and the results from at least three independent
experiments were statistically analyzed by one-way analysis of
variance, using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to represent a statistically significant
difference.
Results
Effect of hyperoside on P. aeruginosa
biofilm formation
Hyperoside is a flavonol glycoside with variety of
biological activities, including antioxidant, anticancer,
antihyperglycemic and anti-inflammatory functions (25–27).
However, prior to the current study, hyperoside has demonstrated
little antibacterial activity (20,28). The
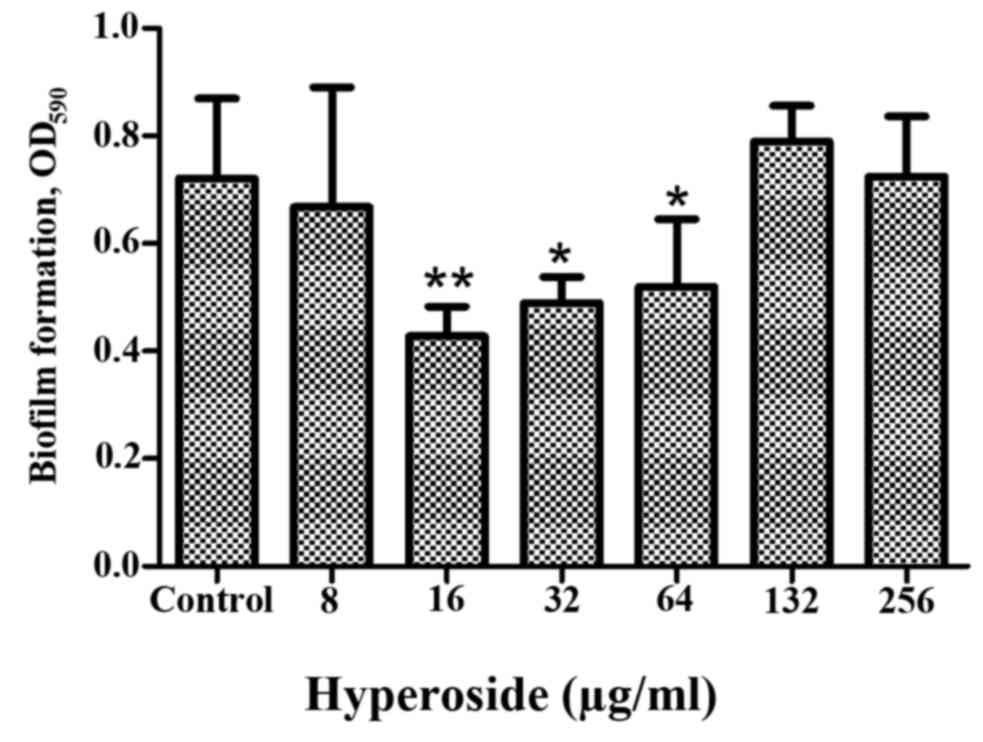
dose-effect experiment of the present study demonstrated that 256
µg/ml hyperoside exhibited no inhibitory effects, and 16 µg/ml
hyperoside had the strongest inhibitory effect on P.
aeruginosa biofilm formation. All doses of hyperoside between
16–64 µg/ml demonstrated biofilm inhibition to an extent (Fig. 1). As hyperoside concentration
decreased, (from 64 to 16 µg/ml) the inhibition rate of biofilm
formation increased. However, the biofilm inhibition activity was
not concentration dependent. A similar mode of action has been
observed for another traditional medicine component, catechins
(29). Although hyperoside does not
exhibit antibacterial activity at experimental concentrations,
higher hyperoside concentrations may have other pharmacological
activities that weaken the effect of biofilm formation.
Effect of hyperoside on growth
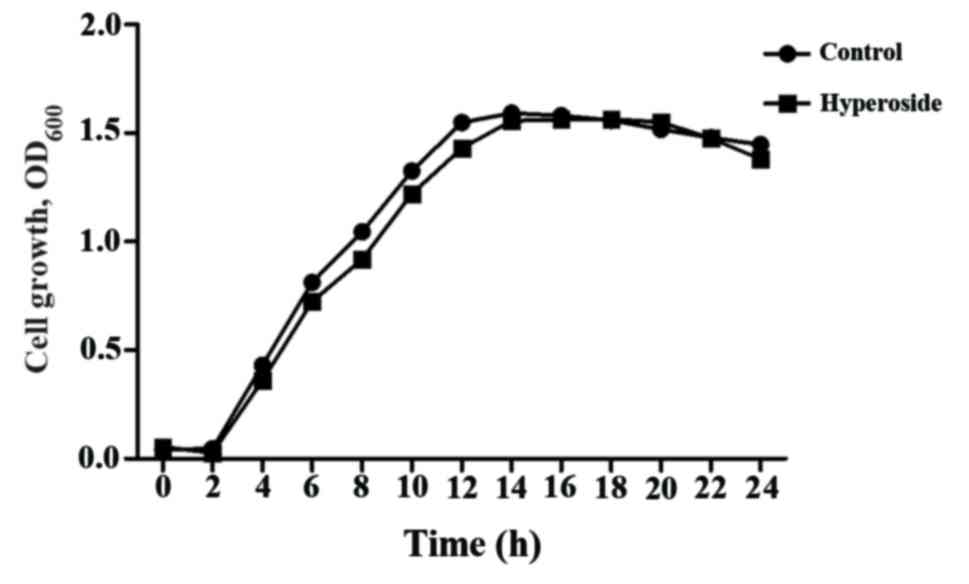
A growth curve using 16 µg/ml hyperoside is
presented (Fig. 2) and the
negligible difference in coincident bacterial growth suggests that
the biofilm inhibition effect was completely unrelated to
antibacterial activity.
CLSM observation
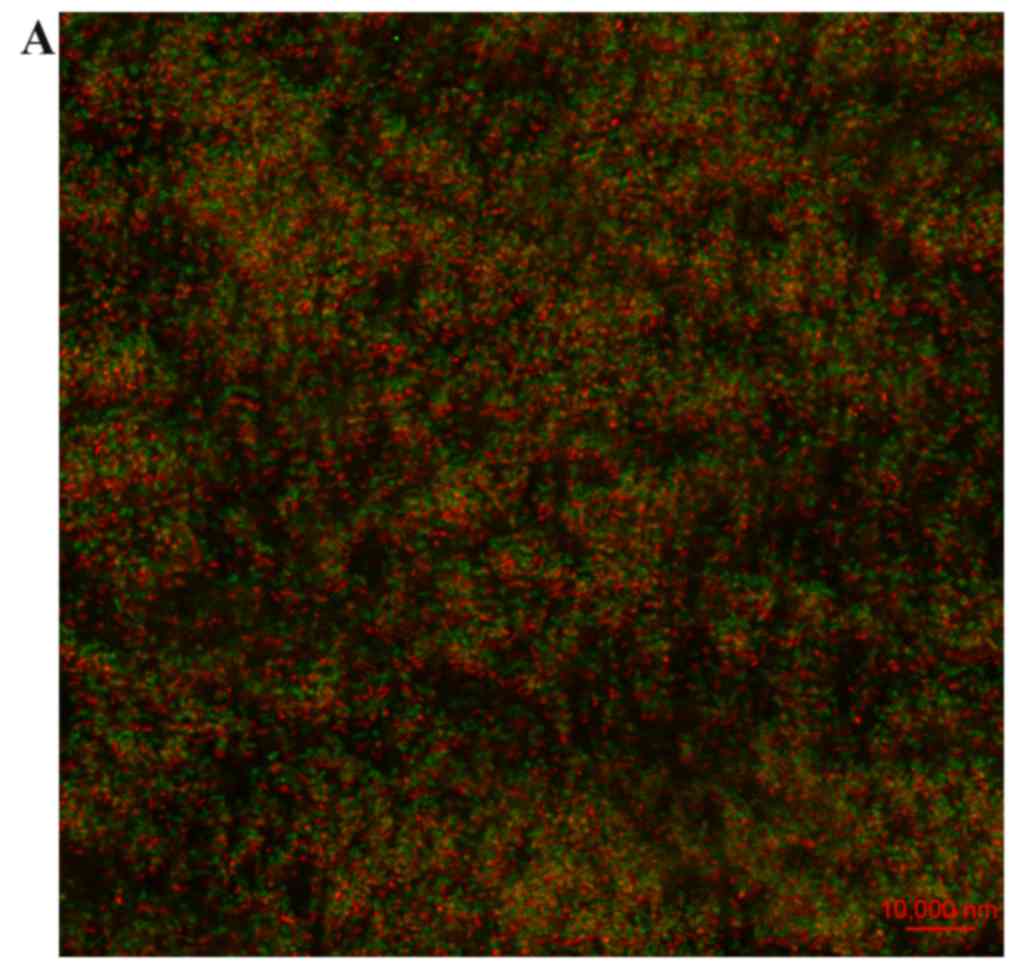
The inhibitory effect of 16 µg/ml hyperoside was
confirmed by CLSM micrographs of the P. aeruginosa biofilm.
Figure 3 shows that the biofilm of
the hyperoside group was sparse (Fig.
3B) compared with the control group (Fig. 3A) and the amount of bacteria and
polysaccharide was clearly decreased.
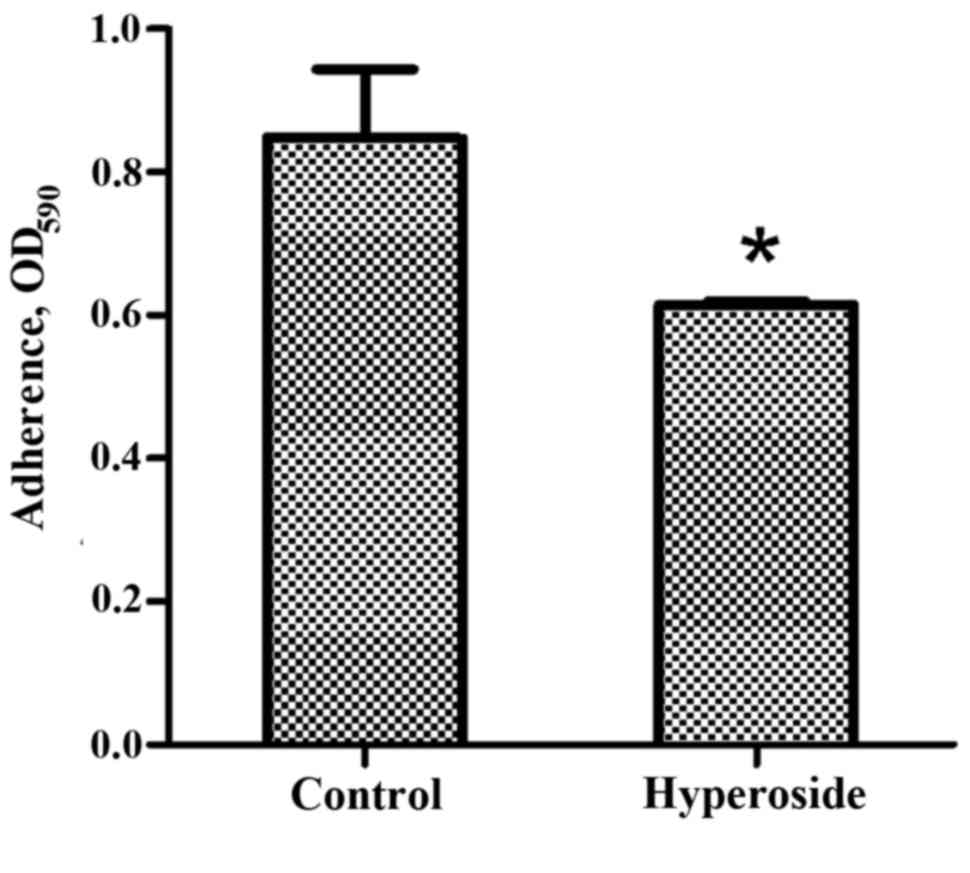
Effect of hyperoside on adhesion
Hyperoside had a significant inhibitory effect on
PAO1 cell adhesion, a process involved in initial biofilm formation
(P<0.05; Fig. 4). The inhibition
of initial adherence by hyperoside suggests that it may decrease
and delay new biofilm formation.
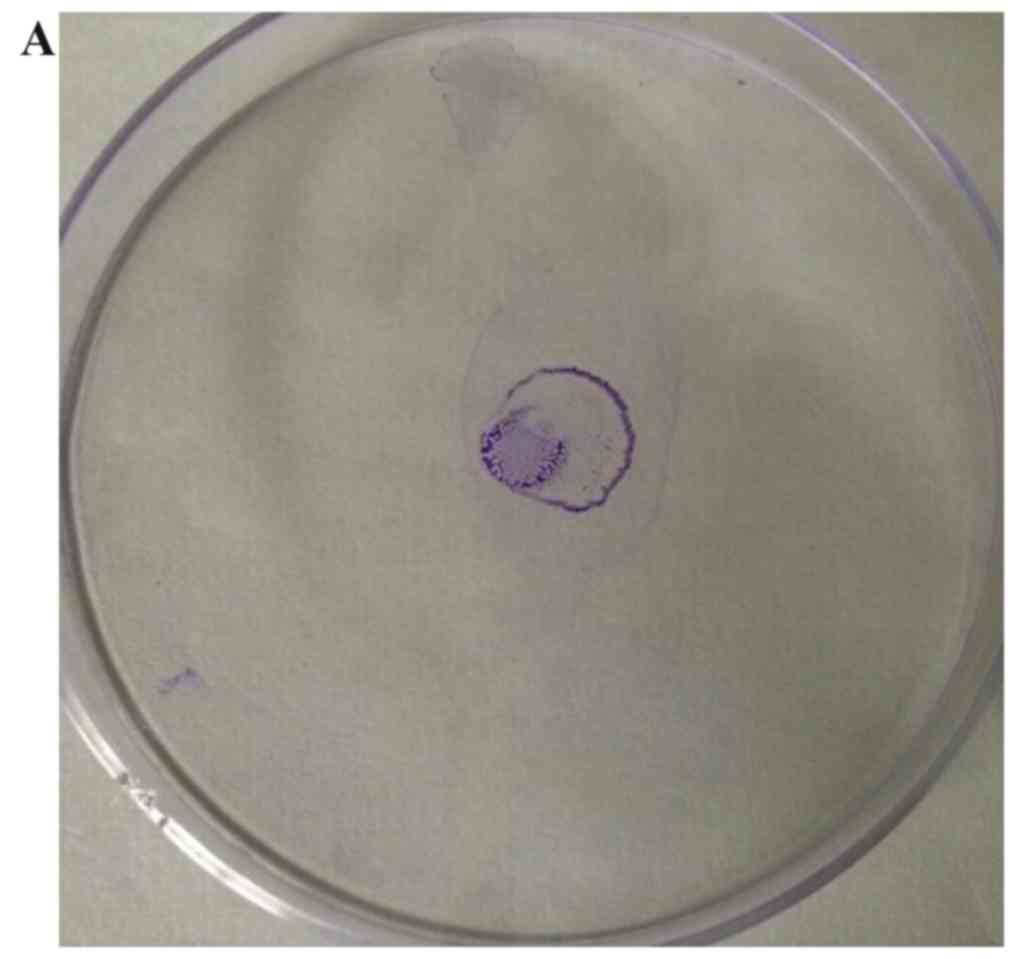
Effect of hyperoside on motility
Twitching motility is mediated by type 4 pili
(30) and this was inhibited by
hyperoside (Fig. 5). Accordingly,
twitching motility may be decreased by inhibiting the activity of
type 4 pili. The movement of pili is the first stage of biofilm
formation. As an essential step for irreversible adhesion, it can
affect the morphology and structure of the biofilm. Therefore,
hyperoside may inhibit the biofilm formation of P.
aeruginosa through inhibiting its type 4 pili.
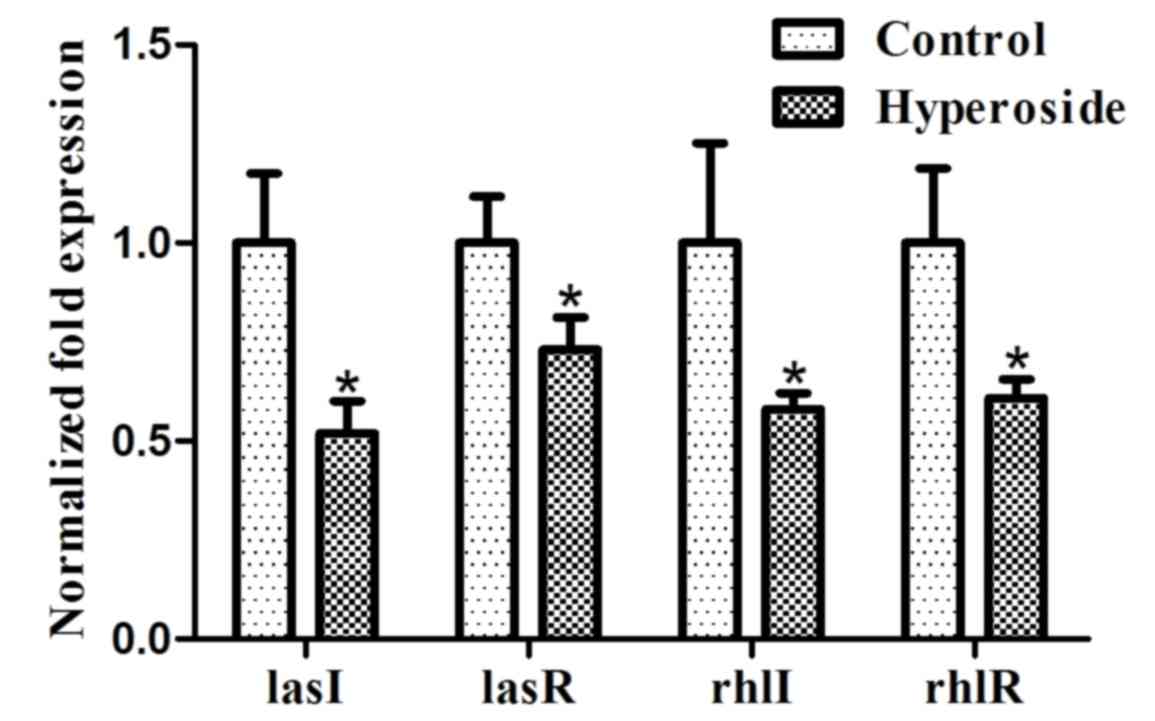
Effect of hyperoside on gene
expression
P. aeruginosa has two well-studied quorum
sensing (QS) systems, las and rhl. The las system is comprised of
the transcriptional activator LasR and the autoinducer (AI)
synthase LasI. The rhl system is comprised of the transcriptional
regulatory protein RhlR and the RhlI AI synthase (31). The las and rhl systems are important
for P. aeruginosa biofilm development (32,33).
Hyperoside inhibits a multitude of factors involved in biofilm
formation; therefore, it may inhibit the QS systems. RT-qPCR was
used to detect lasR, lasI, rhlR and rhlI gene expression. The
results from RT-qPCR indicated that hyperoside inhibited the
expression of the lasR, lasI, rhlR and rhlI genes as the
downregulation of transcription in these genes was significant
(P<0.05; Fig. 6). Similar effects
have been observed regarding other flavonol glycosides in previous
studies (34).
Discussion
Bacteria living in biofilms cause 80% of bacterial
infections (35). Due to the high
antibiotic resistance of biofilms, novel drugs are required to
treat biofilm infections. Previous studies have shown that the
formation of biofilm can be inhibited with sub-minimum inhibitory
concentrations of certain antibiotics, such as imipenem and
erythromycin (36,37). However, the long-term use of
antibiotics will give rise to drug-resistant bacteria. An
increasing number of studies have found that natural products,
including baicalin, allicin and garlicin, exhibit an inhibitory
effect on biofilms (38,39). The use of them for biofilm inhibition
has been successful and has the potential to identify novel
medicines. Currently, there is no natural drug permitted for use in
the clinical treatment of biofilm infection. In this study, we
demonstrated that hyperoside treatment of PAO1 attenuated biofilm
formation, which is related to the QS system. Twitching motility is
a type IV pili-driven movement by which bacteria can adhere and
spread biofilm over a surface (40,41).
Type IV pili is also controlled by QS in P. aeruginosa
(42). In the current study, it was
found that hyperoside-treated PAO1 exhibited reduced adherence, as
well as twitching motility, suggesting that hyperoside impairs the
QS system to inhibit PAO1 adherence, motility and biofilm
formation. Additionally, quantitative analysis of gene expression
showed that hyperoside inhibited the expression of lasR, lasI, rhlR
and rhlI genes involved in the QS system, which indicates that
hyperoside affected P. aeruginosa biofilm formation by
repressing the activity of las and rhl systems.
In view of its structure and function, biofilm is
difficult to be removed completely by a single drug treatment.
Therefore, drug combinations are often used to overcome biofilm
formation, especially antibiotic-antibiotic combinations (43–45).
Despite a favorable anti-biofilm effect, the combination of
antibiotics can result in the generation of drug resistant
bacteria, and enhance the toxicity to humans. Several studies have
showed that phytochemicals can potentiate the activity of
eradicating biofilm of antibiotics in combination (46–49). The
combinations of antibiotic sulfamethoxazole with protocatechuic
acid/ellagic acid/gallic acid and tetracycline with gallic acid,
cefoperazone with allicin showed synergistic mode of interaction
and were highly effective in inhibition P. aeruginosa
biofilm under in vitro conditions (50). Vitexin has been found to potentiate
the anti-biofilm activity of azithromycin and gentamicin on P.
aeruginosa (51). As a type of
nature product, whether hyperoside also has synergistic effects
with antibiotics that inhibit P. aeruginosa biofilm
formation requires further study. Although hyperoside exhibits
antibacterial properties against P. aeruginosa in vitro, its
anti-bacterial effects against P. aeruginosa in vivo are
largely unknown. Defined clinical trials should be conducted to
confirm its efficacy. Furthermore, little is known regarding the
mechanism of hyperoside, and thus warrants further investigation.
Overall, the effect and mechanisms of hyperoside require further
assessment, including analysis using in vivo techniques.
Acknowledgements
The current study was supported by the Application
Development Project of Chongqing (grant no.
cstc2014yykfA110021).
References
|
1
|
Gristina AG, Oga M, Webb LX and Hobgood
CD: Adherent bacterial colonization in the pathogenesis of
osteomyelitis. Science. 228:990–993. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goossens H: Susceptibility of
multi-drug-resistant Pseudomonas aeruginosa in intensive care
units: Results from the European MYSTIC study group. Clin Microbiol
Infect. 9:980–983. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chatterjee M, Anju CP, Biswas L, Kumar V
Anil, Mohan C Gopi and Biswas R: Antibiotic resistance in
Pseudomonas aeruginosa and alternative therapeutic options. Int J
Med Microbiol. 306:48–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Musken M, Di Fiore S, Romling U and
Häussler S: A 96-well-plate-based optical method for the
quantitative and qualitative evaluation of Pseudomonas aeruginosa
biofilm formation and its application to susceptibility testing.
Nat Protoc. 5:1460–1469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Høiby N, Johansen H Krogh, Moser C, Song
Z, Ciofu O and Kharazmi A: Pseudomonas aeruginosa and the in vitro
and in vivo biofilm mode of growth. Microbes Infect. 3:23–35. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ceri H, Olson ME, Stremick C, Read RR,
Morck D and Buret A: The Calgary Biofilm Device: New technology for
rapid determination of antibiotic susceptibilities of bacterial
biofilms. J Clin Microbiol. 37:1771–1776. 1999.PubMed/NCBI
|
|
7
|
Anwar H and Costerton JW: Enhanced
activity of combination of tobramycin and piperacillin for
eradication of sessile biofilm cells of Pseudomonas aeruginosa.
Antimicrob Agents Chemother. 34:1666–1671. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Delden C and Iglewski BH: Cell-to-cell
signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis.
4:551–560. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Wang T, Hu J, Ren C, Lei H, Hou Y
and Brantner AH: Anti-biofilm activity of TanReQing, a Traditional
Chinese Medicine used for the treatment of acute pneumonia. J
Ethnopharmacol. 134:165–170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Palombo EA: Traditional medicinal plant
extracts and natural products with activity against oral bacteria:
Potential application in the prevention and treatment of oral
diseases. Evid Based Complement Alternat Med. 2011:6803542011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sayem SM, Manzo E, Ciavatta L, Tramice A,
Cordone A, Zanfardino A, De Felice M and Varcamonti M: Anti-biofilm
activity of an exopolysaccharide from a sponge-associated strain of
Bacillus licheniformis. Microb Cell Fact. 10:742011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Shang F, Meng Y, Li L, Cui Y,
Zhang M, Qi K and Xue T: Ethanol extract of Sanguisorba officinalis
L. inhibits biofilm formation of methicillin-resistant
Staphylococcusaureus in an ica-dependent manner. J Dairy Sci.
98:8486–8491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren S, Wu M, Guo J, Zhang W, Liu X, Sun L,
Holyst R, Hou S, Fang Y and Feng X: Sterilization of
polydimethylsiloxane surface with Chinese herb extract: A new
antibiotic mechanism of chlorogenic acid. Sci Rep. 5:104642015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen ZW, Ma CG and Xu SY: Mechanism of
analgesic action of hyperin. Yao Xue Xue Bao. 24:326–330. 1989.(In
Chinese). PubMed/NCBI
|
|
15
|
Xin Q and Chen S: Advances in the study of
the protection of hyperin against ischemic injuries of tissues and
organs. Zhong Yao Cai. 26:213–215. 2003.(In Chinese). PubMed/NCBI
|
|
16
|
Wang WQ, Ma CG and Xu SY: Protective
effect of hyperin against myocardial ischemia and reperfusion
injury. Zhongguo Yao Li Xue Bao. 17:341–344. 1996.PubMed/NCBI
|
|
17
|
Verma N, Amresh G, Sahu PK, Mishra N, Rao
ChV and Singh AP: Pharmacological evaluation of hyperin for
antihyperglycemic activity and effect on lipid profile in diabetic
rats. Indian J Exp Biol. 51:65–72. 2013.PubMed/NCBI
|
|
18
|
Lee S, Park HS, Notsu Y, Ban HS, Kim YP,
Ishihara K, Hirasawa N, Jung SH, Lee YS, Lim SS, et al: Effects of
hyperin, isoquercitrin and quercetin on lipopolysaccharide-induced
nitrite production in rat peritoneal macrophages. Phytother Res.
22:1552–1556. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marčetić MD, Milenković MT, Lakušić DV and
Lakušić BS: Chemical Composition and antimicrobial activity of the
essential oil and methanol extract of Hypericum aegypticum subsp.
webbii (Spach) N. Robson. Chem Biodivers. 13:427–436. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee S, Shin DS, Oh KB and Shin KH:
Antibacterial compounds from the leaves of Acanthopanax senticosus.
Arch Pharm Res. 26:40–42. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang SS, Wang DM, Pu WJ and Li DW:
Phytochemical profiles, antioxidant and antimicrobial activities of
three Potentilla species. BMC Complement Altern Med. 13:3212013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Neidig A, Yeung AT, Rosay T, Tettmann B,
Strempel N, Rueger M, Lesouhaitier O and Overhage J: TypA is
involved in virulence, antimicrobial resistance and biofilm
formation in Pseudomonas aeruginosa. BMC Microbiol. 13:772013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bala A, Kumar R and Harjai K: Inhibition
of quorum sensing in Pseudomonas aeruginosa by azithromycin and its
effectiveness in urinary tract infections. J Med Microbiol.
60:300–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xing HY, Liu Y, Chen JH, Sun FJ, Shi HQ
and Xia PY: Hyperoside attenuates hydrogen peroxide-induced L02
cell damage via MAPK-dependent Keap-Nrf-ARE signaling pathway.
Biochem Biophys Commun. 410:759–765. 2011. View Article : Google Scholar
|
|
26
|
Li W, Liu M, Xu YF, Feng Y, Che JP, Wang
GC and Zheng JH: Combination of quercetin and hyperoside has
anticancer effects on renal cancer cells through inhibition of
oncogenic microRNA-27a. Oncol Rep. 31:117–124. 2014.PubMed/NCBI
|
|
27
|
Ku SK, Kwak S, Kwon OJ and Bae JS:
Hyperoside inhibits high-glucose-induced vascular inflammation in
vitro and in vivo. Inflammation. 37:1389–1400. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abedini A, Roumy V, Mahieux S, Biabiany M,
Standaert-Vitse A, Rivière C, Sahpaz S, Bailleul F, Neut C and
Hennebelle T: Rosmarinic acid and its methyl ester as antimicrobial
components of the hydromethanolic extract of Hyptis atrorubens
poit. (Lamiaceae). Evid Based Complement Alternat Med.
2013:6045362013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsunaga T, Nakahara A, Minnatul KM,
Noiri Y, Ebisu S, Kato A and Azakami H: The inhibitory effects of
catechins on biofilm formation by the periodontopathogenic
bacterium, Eikenella corrodens. Biosci Biotechnol Biochem.
74:2445–2450. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giltner CL, van Schaik EJ, Audette GF, Kao
D, Hodges RS, Hassett DJ and Irvin RT: The Pseudomonas aeruginosa
type IV pilin receptor binding domain functions as an adhesin for
both biotic and abiotic surfaces. Mol Microbiol. 59:1083–1096.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Davies DG, Parsek MR, Pearson JP, Iglewski
BH, Costerton JW and Greenberg EP: The involvement of cell-to-cell
signals in the development of a bacterial biofilm. Science.
280:295–298. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morici LA, Carterson AJ, Wagner VE, Frisk
A, Schurr JR, Hönerzu Bentrup K, Hassett DJ, Iglewski BH, Sauer K
and Schurr MJ: Pseudomonas aeruginosa AlgR represses the Rhl
quorum-sensing system in a biofilm-specific manner. J Bacteriol.
189:7752–7764. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Kievit TR, Gillis R, Marx S, Brown C
and Iglewski BH: Quorum-sensing genes in Pseudomonas aeruginosa
biofilms: Their role and expression patterns. Appl Environ
Microbiol. 67:1865–1873. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng Z, Qian L, Cao L, Tan H, Huang Y, Xue
X, Shen Y and Zhou S: Virtual screening for novel quorum sensing
inhibitors to eradicate biofilm formation of Pseudomonas
aeruginosa. Appl Microbiol Biotechnol. 79:119–126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vo GD, Brindle E and Heys J: An
experimentally validated immersed boundary model of fluid-biofilm
interaction. Water Sci Technol. 61:3033–3040. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cirioni O, Silvestri C, Ghiselli R, Kamysz
W, Minardi D, Castelli P, Orlando F, Kamysz E, Provinciali M,
Muzzonigro G, et al: In vitro and in vivo effects of sub-MICs of
pexiganan and imipenem of Pseudomonas aeruginosa adhesion and
biofilm development. Infez Med. 21:287–295. 2013.PubMed/NCBI
|
|
37
|
Zhao YL, Zhou YH, Chen JQ, Huang QY, Han
Q, Liu B, Cheng GD and Li YH: Quantitative proteomic analysis of
sub-MIC erythromycin inhibiting biofilm formation of S, suis in
vitro. J Proteomics. 116:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang C, Cheng H, Zhang X, Xu S, Guan Y, Yu
L and Yun Y: In vitro activity of baicalin against non-albicans
Candida biofilms. Zhongguo Zhong Yao Za Zhi. 35:639–641. 2010.(In
Chinese). PubMed/NCBI
|
|
39
|
Saleem M, Nazir M, Ali MS, Hussain H, Lee
YS, Riaz N and Jabbar A: Antimicrobial natural products: An update
on future antibiotic drug candidates. Nat Prod Rep. 27:238–254.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tolker-Nielsen T, Brinch UC, Ragas PC,
Andersen JB, Jacobsen CS and Molin S: Development and dynamics of
Pseudomonas aeruginosa sp. biofilms. J Bacteriol. 182:6482–6489.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sauer K, Camper AK, Ehrlich GD, Costerton
JW and Davies DG: Pseudomonas aeruginosa displays multiple
phenotypes during development as a biofilm. J Bacteriol.
184:1140–1154. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Köhler T, Curty LK, Barja F, van Delden C
and Pechére JC: Swarming of Pseudomonas aeruginosa is dependent on
cell-to-cell signaling and requires flagella and pili. J Bacteriol.
182:5990–5996. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Olson KM, Starks CM, Williams RB,
O'Neil-Johnson M, Huang Z, Ellis M, Reilly JE and Eldridge GR:
Novel pentadecenyl tetrazole enhances susceptibility of
methincillin-resistant Staphylococcus aureus biofilm to gentamicin.
Antimicrob Agents Chemother. 55:3691–3695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pettit RK, Weber CA, Lawrence SB, Pettit
GR, Kean MJ and Cage GD: In vivo activity of anprocide alone and in
vitro activity in combination with conventional antibiotics against
Staphylococcus aureus and Staphylococcus epidermidis biofilms. J
Med Microbiol. 58:1203–1206. 2010. View Article : Google Scholar
|
|
45
|
McConeghy KW and LaPlante KL: In vitro
activity of tigecycline in combination with gentamicin against
biofilm-forming Staphylococcus aureus. Diagn Microbiol Infect Dis.
68:1–6. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen YQ, Zuo XJ, Zhu LN, Song ZJ, Shi HZ,
Zuo P and Guo XH: In vitro effects of honeysuckle aqueous extracts
alone and in combination with ceftazidime on Pseudomonas aeruginosa
biofilms. Chin J Microbiol Immunol. 24:738–742. 2004.
|
|
47
|
Kong JL, Liu XL, Chen YQ, et al: In vitro
effect of Scutellaria aqueous extracts combination with
levofloxacin on Pseudomonas aeruginosa biofilm. Tianjin Med J.
36:331–333. 2008.
|
|
48
|
Huang XM, Huang LC, Fang ZH, et al:
Synergism of Sophora flavescens ait and ciprofloxacin on
Pseudomonas aeruginosa biofilms. J Shaoguan Univ. 27:60–63.
2006.
|
|
49
|
Zhou Q, Deng CH, Zhang W, et al: In vitro
effects of aqueous extract of Sophora in combination with
ceftazidime on elimination of Pseudomonas aeruginosa biofilms. J
New Chin Med. 40:98–99. 2008.
|
|
50
|
Jayaraman P, Sakharkar MK, Lim CS, Tang TH
and Sakharkar KR: Activity and interactions of antibiotic and
phytochemical combinations against Pseudomonas aeruginosa in vitro.
Int J Biol Sci. 6:556–568. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Das MC, Sandhu P, Gupta P, Rudrapaul P, De
UC, Tribedi P, Akhter Y and Bhattacharjee S: Attenuation of
Pseudomonas aeruginosa biofilm formation by Vitexin: A
combinatorial study with azithromycin and gentamicin. Sci Rep.
6:233372016. View Article : Google Scholar : PubMed/NCBI
|