Introduction
Gestational diabetes mellitus (GDM) is a common
complication of pregnancy associated with an increased incidence of
pregnancy complications, adverse pregnancy outcomes, and maternal
and fetal risks of chronic health conditions later in life
(1). The occurrence of GDM is a
multi-factor and multi-stage process, which is directed by the
environmental, dietary or genetic factors. However, the specific
mechanism has not been fully elucidated. Well-documented risk
factors for GDM include pre-pregnancy overweight and obesity
(2). But some reports also showed
that only a few people suffered from GDM under the context of the
same diet, suggesting that different individuals displayed
different susceptibility to food habits (3). Several studies referred to the close
association between genetic variants and the risk of GDM (4).
Single nucleotide polymorphisms (SNPs) are the major
type of genetic variation in human beings. Many studies reported
that allelic variations of SNPs can influence the expression or
function of their hosting genes. Increasing evidence revealed that
microRNA (miRNA)-related SNPs significantly altered the biogenesis
and/or function of the corresponding miRNA (5). miRNA is a small non-coding
single-stranded RNA consisting of 18–24 nucleotides, widespread
throughout of human genome (6). They
typically regulate genes by binding to the 3′ UTR of the target
mRNA, which destabilizes the mRNA and prevents translation
(7). miRNAs are emerging as agents
of metabolic and malignant regulation in development and disease.
Lethal-7 (let-7) miRNA has gained notoriety owing to its regulation
of stem cell differentiation and essential role in normal
development (8). Let-7 miRNA
cooperates with multiple transcription factors and directly
regulates the targeted gene expression. A RNA binding protein and
stem cell marker Lin28 is a main regulator of let-7, and is
initially found in C. elegans to regulate the fate of cell
both in early development and terminal differentiation (9). There is a double-negative feedback loop
between Lin28 and let-7: let-7 can repress the post-transcriptional
translation of Lin28 (8), while
Lin28 blocks the maturation of let-7 miRNA (10).
Meta-analysis of published data confirmed that let-7
targets are enriched for gene containing SNPs associated with type
2 diabetes mellitus (T2DM) and control of fasting glucose in humans
(11). A SNP located near the let-7
binding site in Lin28, the T/C variants of rs3811463 could lead to
differential regulation of Lin28 by let-7 (12). The rs3811463 polymorphism in the
let-7/Lin28 pathway significantly increased the risk of T2DM
(13). Insulin resistance is the
major determinant of T2DM and GDM (14). So there might be some relationship
between genetic polymorphisms in the let/Lin28 pathway and the risk
of GDM.
In our study, we constructed GDM rat model via high
fat diet and STZ injection and then examined the body weight and
blood glucose level. The primary skeletal muscle cells were
successfully incubated and transfected with T/C variants of
rs3811463 and then glucose uptake and insulin sensitivity were
examined. Furthermore, the glucose metabolism-related and insulin
sensitivity-related genes were detected after various recombined
vectors transfection. Taken together, we found that C allele of
rs3811463 mitigated the expression of let-7, regulated the
expression of glucose metabolism-related and insulin
sensitivity-related genes to participate in the development of
GDM.
Materials and methods
Construction of GDM rats model
Total 40 adult SPF Wistar rats, weighing 190–210 g,
7–8 weeks old, were purchased from Shanghai Silaike Laboratory
Animal Limited Liability Company (Shanghai, China). Rats were kept
at standard laboratory conditions at 18–25°C with 12 h light: 12 h
dark. Standard rodent feed and water were available ad libitum. All
procedures were performed in accordance with the Guide for the
Humane Use and Care of Laboratory. This study was approved by the
Committees of The First Affiliated Hospital with Nanjing Medical
University and Jiangsu Women and Children Health Hospital. After a
week of adaptation, the female and male rats were mated at the
ratio of 2:1. Next morning, the female vaginal suppository was
checked and the pregnant rats were housed individually. The
pregnant rats were fed with a high fat diet (HFD, 58% fat, 25%
protein and 17% carbohydrate, as a percentage of total kcal) for an
initial period of two weeks (15).
Thereafter, rats received a 60 mg/kg intraperitoneal injection of
streptozotocin (STZ) in freshly prepared sodium citrate buffer (0.1
mM, pH 4.5). The normal control (NC) group was administrated with
the same amount of sodium citrate buffer. Rats with blood glucose
levels after 72 h of STZ injection higher than 16.67 mM were
diagnosed as GDM for further experiments.
Isolation of primary cells
STZ-induced diabetic female rats were anaesthetizes
with injection of pentobarbital (60 mg/kg) into abdominal cavity.
The musculi quadriceps femoris of hind legs were isolated and
washed with sterile PBS buffer to remove the connective tissues.
Then the isolated tissues were sheared and digested with
collagenase IA (Sigma, USA) at 4°C overnight. After centrifugation,
skeletal muscle cells were cultured with Dulbecco modified Eagle
medium (DMEM, GIBCO, USA) supplemented with 10% fetal bovine serum
(FBS, GIBCO, USA) and 1% myllicin in a humidified atmosphere of 5%
CO2 at 37°C.
Recombined vectors construction
Genomic DNA was extracted from the primary skeletal
muscle cells using a DNA extraction Kit (Invitrogen, USA). An 885
bp fragment of the 3′ UTR of rat Lin28 gene centering rs3811463 and
the predicted complementary site of let-7 were inserted into
pEF6/V5-His-TOPO vector. A mutant plasmid with C allele at the site
of rs3811463 was constructed using a site-directed mutagenesis kit
(Stratagene, La Jolla, CA, USA). All recombined plasmids were
verified by direct DNA sequencing. The wild type recombinant and
mutated vectors were named pEF6/V5-His-TOPO: rs3811463T and
pEF6/V5-His-TOPO: rs3811463C, respectively.
Cell transfection
The third passage skeletal muscle cells were seeded
into 6-well plates at the concentration of 1–2×106
cell/well. When the density reached 80–90%, cell transfection was
performed according to the manufacturer's instructions. About 0.8
µg plasmid DNA or 2 µl Lipofectamine 3000 was diluted with 50 µl
Opti-MEM FBS-free DMEM, respectively. After 5 min at room
temperature, the diluted DNA and Lipofectamine 3000 were mixed
gently. About 20 min later at room temperature, about 100 µl of
mixture was added into the cells and cultured at 37°C incubators.
After 6 h, the medium was replaced with fresh DMEM containing 10%
FBS.
Glucose uptake and insulin sensitivity
assay
Muscle cells were pretreated in low-glucose DMEM for
12 h, and then transfected with pEF6/V5-His-TOPO, pEF6/V5-His-TOPO:
rs3811463T or pEF6/V5-His-TOPO: rs3811463C, respectively. After 48
h, the old medium was removed, and cells were incubated with 4 ml
of HEPES-buffer saline containing [3H]-2-deoxy-D-glucose
(1.0 mCi/ml) for 30 min. After being washed with PBS, cells were
trypsinized with 0.25% trypsin (Sigma, USA). The radioactivity was
determined by scintillation counting. All experiments were assayed
in at least three different independent repeat. For insulin
sensitivity assay, cells were pretreated with 10 µg/ml of insulin
for 4 h, and then glucose uptake was detected according to the
above protocol.
Quantitative reverse transcription-PCR
(qRT-PCR)
Total RNA was extracted from muscle cells with
Trizol reagent (Sigma, USA) in accordance with the manufacturer's
instructions. The concentration and quality was measured by
Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA).
About 1 µg of total RNA was reverse transcribed with
SuperScript® III First Strand kit (Invitrogen, USA)
according to the manufacturer's protocol. QRT-PCR was performed
using TaqManGene Expression MasterMix (Thermo, USA). The parameters
for qRT-PCR assay were as follows: 95°C for 10 min, 40 cycles of
95°C for 15 sec, 60°C for 1 min. QRT-PCR was performed in
triplicate with Applied Biosystems 7500 PCR System (Applied
Biosystems, USA) according to the standard protocol. The mRNA
levels of IRS1, IGF2BP2 and GCK genes were normalized to the level
of GAPDH using the method of 2−ΔΔCT and were represented
as fold induction. The primer sequences for qRT-PCR are listed in
Table I.
 | Table I.Primers for qRT-PCR. |
Table I.
Primers for qRT-PCR.
| Genes | Forward primers
(5′-3′) | Reverse primers
(5′-3′) |
|---|
| IRS1 |
cgatggcttctcagacgtg |
cagcccgcttgttgatgttg |
| IGF2BP2 |
gactaccccgaccagaactg |
gaggcgggatgttccgaatc |
| GCK |
accaagcggtatcagcatgtg |
tggacttctctgtgattggca |
| GAPDH |
tgtgggcatcaatggatttgg |
acaccatgtattccgggtcaat |
Western blot analysis
After various treatments, muscle cells were
harvested and lysed with RIPA buffer (Sigma, USA). The
concentration of protein was detected with bicinchoninic acid kit
(Pierce, Rockford, IL). Proteins (40 µg/lane) were subjected into
12% SDS-polyacrylamide gel and then transferred onto a
polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA,
USA). Subsequently, the membrane was blocked with 2% non-fat milk
in TBS containing 0.05% Tween-20 (TBST) for 1 h at room temperature
and incubated overnight at 4°C with the primary antibodies against
Sirtuin 1 (SIRT1, Abcam, USA). After being washed with TBST for
five times, the membranes were probed with horseradish
peroxidase-conjugated secondary antibodies (1:10,000 dilution,
zhongshan, Beijing, China). The membranes were developed with the
enhanced chemiluminescence kit (Amersham). Relative band intensity
of SIRT1 was detected with Quantity One v4.62 (Bio-Rad, USA) and
normalized to GAPDH.
Statistical analysis
Statistical analysis in the present study was
performed with Graphpad prism 6.0 (GraphPad Software, La Jolla, CA,
USA). All data from the experiments are presented as the mean ±
standard deviation. We used one-way analysis of variance (ANOVA) to
determine significance between two groups via SPSS software
(version 16.0, SPSS Inc, Chicago, Illinois). The letters in the
Figures indicate the significant difference (P<0.05) in various
treatments by one-way ANOVA.
Results
GDM rat model was successfully
constructed by high fat diet and STZ injection
In the experiment, we selected 13 of pregnant female
rats for model construction. To verify the effect of high fat diet
and STZ injection, we examined the body weight and blood glucose of
tail vein at the 3rd, 6th, 9th and 12th d after STZ injection shown
in Table II. There was no
significant difference on the body weight at the 3rd and 6th d
between NC and GDM groups (P>0.05). However, at the 9th and 12th
d, the body weight of GDM was obviously lower than that in NC group
(P<0.05). Meanwhile, the blood glucose in GDM group was
significantly higher than that in NC group at the 3rd, 6th, 9th and
12th d (P<0.05). Thus we concluded that GDM rat model was
successfully constructed for follow-up experiments.
 | Table II.The examination of body weight and
blood glucose level. |
Table II.
The examination of body weight and
blood glucose level.
|
| Time (d) |
|---|
|
|
|
|---|
| Group | 3rd | 6th | 9th | 12th |
|---|
| Body weight (g) |
|
|
|
|
| NC | 171.12±2.6 | 184.95±1.5 | 203.08±5.1 | 234.48±3.4 |
| GDM | 174.00±3.7 | 185.74±0.8 |
180.71±2.8a |
191.58±2.1a |
| Blood glucose
(mM) |
|
|
|
|
| NC | 17.69±0.9 | 17.26±1.9 | 17.55±3.3 | 17.39±1.7 |
| GDM |
21.17±1.4a |
27.59±2.9a |
35.35±0.7a |
40.51±0.8a |
The effect of replacement of the T
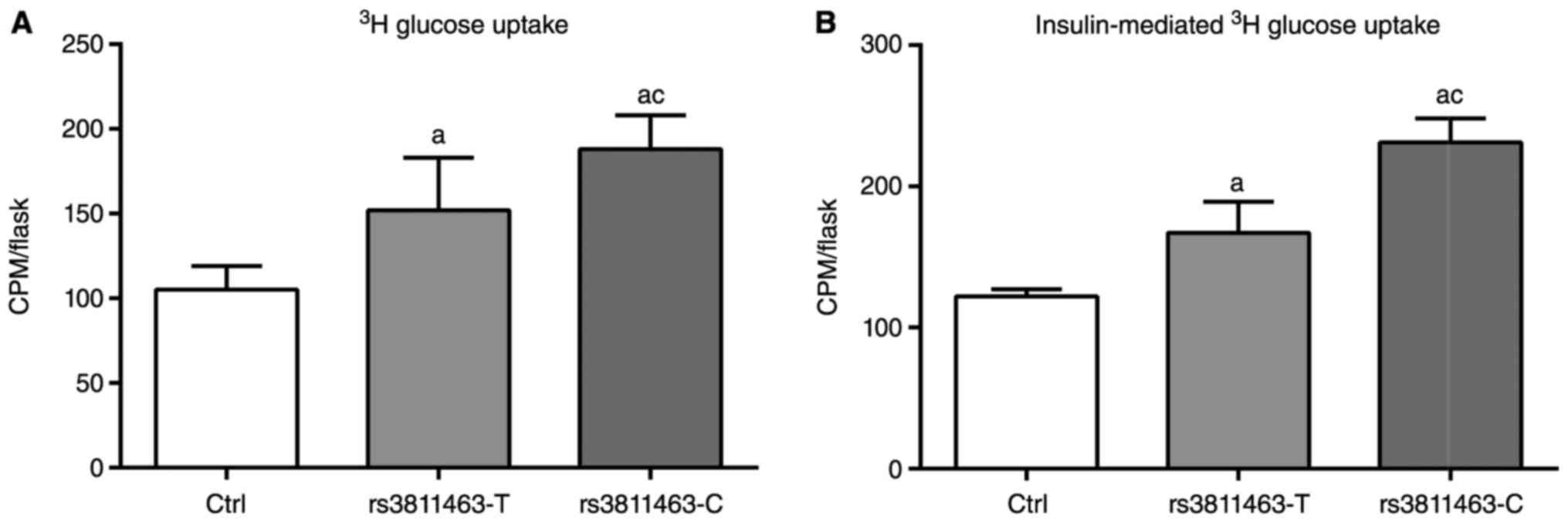
allele by the C allele at rs3811463 on glucose uptak
To investigate the effect of replacement of the T
allele by the C allele at rs3811463 on glucose uptake, we
transfected muscle cells with the wild and mutated vectors and
performed 3H glucose uptake experiments in Fig. 1A. In pEF6/V5-His-TOPO-transfected
cells (Ctrl group), the radioactivity of lipid extraction (132±14
CPM/flask) was lower than that in rs3811463-T (152±31 CPM/flask)
and rs3811463-C (188±20 CPM/flask) groups (P<0.05). And the
radioactivity of lipid extraction after rs3811463-C transfection
was higher than that in rs3811463-T-transfected group (P<0.05).
These data showed that the overexpression of Lin28 enhanced the
activity of glucose uptake, and the replacement of the T allele by
the C at rs3811463 promoted the glucose uptake.
To test the insulin sensitivity, cells were
pretreated with insulin for 4 h, and then transfected with various
vectors for glucose uptake detection. In Fig. 1B, the radioactivity of lipid
extraction in pEF6/V5-His-TOPO-transfected cells (Ctrl group) was
147±5 CPM/flask, which was significantly lower than that in
rs3811463-T (167±22 CPM/flask) and rs3811463-C (215±17 CPM/flask)
groups (P<0.05). What's more, rs3811463-C transfection increased
the radioactivity than rs3811463-T. These results showed that the
overexpression of Lin28 enhanced the activity of glucose uptake
after insulin stimulation, and the replacement of the T allele by
the C at rs3811463 promoted the glucose uptake after insulin
stimulation.
C allele at rs3811463 affected the
expression of IGF2BP2 and GCK
To examine the mechanism by which C allele at
rs3811463 modulated the glucose uptake, we analyzed the expression
of glucose metabolism-related gene by qRT-PCR in Fig. 2. The transfection of rs3811463-T
obviously upregulated the expression of let-7, but had no effect on
the mRNA levels of in glucokinase (GCK), insulin-like growth factor
two binding protein 2 (IGF2BP2) and insulin receptor substrate 1
(IRS1) genes. The rs3811463-C transfection had no effect on the
levels of let-7 and IRS1, but significantly improved the expression
of IG2BP2 and GCK. These data showed that the transfection of
rs3811463 modulated the ability of glucose uptake by affecting the
expression of glucose metabolism-related genes.
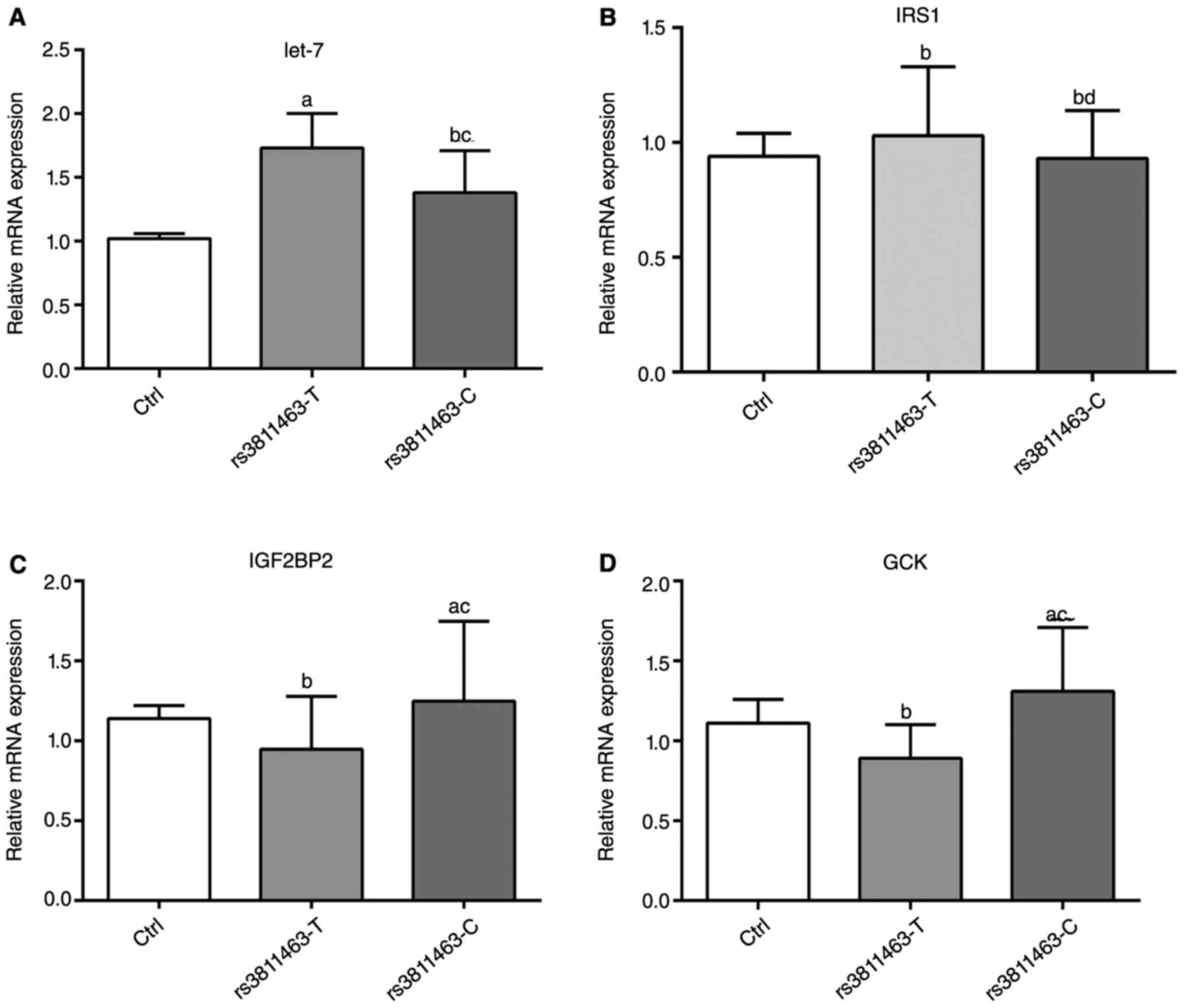 | Figure 2.The mRNA levels of let-7 (A), IRS1
(B), IGF2BP2 (C) and GCK (D) after cell transfection determined by
qRT-PCR. Muscle cells were pretreated in low-glucose DMEM for 12 h,
and then transfected with pEF6/V5-His-TOPO (Ctrl group),
pEF6/V5-His-TOPO: rs3811463T (rs3811463T group) or
pEF6/V5-His-TOPO: rs3811463C (rs3811463C group), respectively.
After 48 h, RNA was isolated for qRT-PCR. GAPDH was used as the
internal control. All experiments were performed in triplicate
independent repeat analyzed with one-way ANOVA. Compared with Ctrl
group, ameans P<0.05, bmeans P>0.05.
Compared with rs3811463T group, cP<0.05,
dP>0.05. |
C allele at rs3811463 modulated the
expression of SIRT1
To explore the reason of rs3811463-T/C transfection
affecting the glucose uptake after insulin stimulation, we examined
the expression of insulin sensitivity-related gene, SIRT1. SIRT1 is
a conserved protein NAD+-dependent deacylases and thus the function
is intrinsically linked to cellular metabolism (16). In Fig.
3, compared with pEF6/V5-His-TOPO-transfected cells (Ctrl
group), the transfection of rs3811463-T and rs3811463-C
significantly enhanced the protein level of SIRT1. But the
rs3811463-C transfection resulted into the higher expression of
SIRT1 than rs3811463-T. These data showed that C allele at
rs3811463 modulated the insulin sensitivity by regulating the
expression of SIRT1.
Discussion
In this study, we investigated the association
between T/C variants of rs3811463 in the let-7/Lin28 pathway and
the risk of GDM. We injected pregnant female rats fed on HFD with
STZ to induce GDM, which is a classic method for GDM induction
(17). STZ injection improved the
body weight and blood glucose, suggesting that GDM model was
successfully constructed for further investigation. The replacement
of the T allele by C allele of rs3811463 affected glucose uptake,
the expression of let-7, glucose metabolism-related and insulin
sensitivity-related genes for GDM. These findings revealed that
genetic variations of rs3811463 may lead to altered status of the
let-7/Lin28 loop and are associated with GDM susceptibility.
Previous studies showed that the highly conserved
miRNA let-7 represses the post-transcriptional translation of
Lin28, and Lin28 in turn blocks the maturation of let-7, forming a
double-negative feedback loop (18).
The Lin28/let-7 axis is highly conserved across the animal kingdom
in organisms as evolutionarily distant as nematode worms and humans
(19). Let-7 family of miRNAs
contributed to the control of glucose homeostasis and insulin
sensitivity, and the knockdown of the let-7 could be taken as a
potential treatment for T2DM (20).
Lin28A and Lin28b transgenic animals are more sensitive to insulin
and have reduced peripheral glucose levels (19), which suggest that let-7/Lin28 axis
regulates glucose metabolism. In our study, we found that the
overexpression of Lin28 improved the ability of glucose uptake and
promoted the expression of let-7. Our data provided new evidence
that let-7/Lin28 pathway was involved in glucose metabolism, which
also provided a new target for DM therapy.
Increasing evidence indicated the association
between genetic variation and the risk of GDM (21). SNPs are the major type of human
genomic variation. Among Han Chinese woman, three SNPs (rs4659441,
rs3811463 and rs6697410) were detected in the 3′UTR of Lin28, and
allelic variation of rs3811463 influenced let-7-related regulation
of Lin28 expression (12). Previous
studies showed that insulin resistance and defects in insulin
secretion play key roles in the etiology of both GDM and T2DM. So
we guess that there might be some link between rs3811463 and GDM
susceptibility. In our study, we found that the replacement of T
allele by C allele significantly modulated the glucose uptake and
insulin sensitivity in GDM model, which revealed that genetic
variations of rs3811463 were associated with the risk of GDM by
disturbing the double-negative feedback loop. This might be the
main reason for the association between gene polymorphism of
let-7/Lin28 and GDM susceptibility. What's more, owing to the
disturbance of the double-negative feedback loop, we found that C
allele of rs3811463 improved the expression of Lin28 to suppress
the level of let-7.
It is indicated some significant associations of GDM
with SNPs in GCK, IGF2BP2 and IRS1 genes (21), which are thought to modulate
pancreatic islet β-cell function (22). In our study, we found that T allele
of rs3811463 had no significant effect on the levels of IRS1,
IGF1BP2 and GCK, however, C allele of rs3811463 improved the
expression of above three genes, suggesting that genetic variation
of rs3811463 was involved in the regulation of glucose-metabolism
related genes. These results revealed the relationship between
genetic variation of rs3811463 and glucose metabolism. SIRT1 is
implicated in the regulation of mitochondrial function, energy
metabolism, and insulin sensitivity (23). In our subject, we observed that T
allele of rs3811463 enhanced the protein level of SIRT1 and the
replacement of T allele by C allele obviously improved the level of
SIRT1. These data showed that let-7/Lin28 pathway regulated insulin
sensitivity by modulating the expression of SIRT1. Taken together,
let-7/Lin28 axis is a key factor for glucose metabolism and insulin
sensitivity by regulating glucose metabolism-related and insulin
sensitivity related genes. These findings revealed that gene
polymorphism of let-7/Lin28 might be a new target for GDM
susceptibility study.
Acknowledgements
The present study was supported by Jiangsu Province,
maternal and child health research projects gynecological science
key projects (F201406).
Glossary
Abbreviations
Abbreviations:
|
GDM
|
gestational diabetes
|
|
SNP
|
single nucleotide polymorphism
|
|
miRNA
|
microRNA
|
|
let-7
|
Lethal-7
|
|
T2DM
|
type 2 diabetes mellitus
|
|
STZ
|
streptozotocin
|
|
qRT-PCR
|
quantitative reverse
transcription-PCR
|
|
GCK
|
glucokinase
|
|
IGF2BP2
|
insulin-like growth factor two binding
protein 2
|
|
IRS1
|
Insulin receptor substrate 1
|
References
|
1
|
Russo LM, Nobles C, Ertel KA, Chasan-Taber
L and Whitcomb BW: Physical activity interventions in pregnancy and
risk of gestational diabetes mellitus: A systematic review and
meta-analysis. Obstet Gynecol. 125:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang C and Ning Y: Effect of dietary and
lifestyle factors on the risk of gestational diabetes: Review of
epidemiologic evidence. Am J Clin Nutr. 94 6 Suppl:1975S–1979S.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bowers K, Tobias DK, Yeung E, Hu FB and
Zhang C: A prospective study of prepregnancy dietary fat intake and
risk of gestational diabetes. Am J Clin Nutr. 95:446–453. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Donin AS, Nightingale CM, Owen CG,
Rudnicka AR, Jebb SA, Ambrosini GL, Stephen AM, Cook DG and Whincup
PH: Dietary energy intake is associated with type 2 diabetes risk
markers in children. Diabetes Care. 37:116–123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nicoloso MS, Sun H, Spizzo R, Kim H,
Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L,
et al: Single-nucleotide polymorphisms inside microRNA target sites
influence tumor susceptibility. Cancer Res. 70:2789–2798. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
8
|
Nguyen LH and Zhu H: Lin28 and let-7 in
cell metabolism and cancer. Transl Pediatr. 4:4–11. 2015.PubMed/NCBI
|
|
9
|
Sun X, Liu J, Xu C, Tang SC and Ren H: The
insights of Let-7 miRNAs in oncogenesis and stem cell potency. J
Cell Mol Med. 20:1779–1788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Viswanathan SR, Daley GQ and Gregory RI:
Selective blockade of microRNA processing by Lin28. Science.
320:97–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu H, Shyh-Chang N, Segrè AV, Shinoda G,
Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG,
et al: The Lin28/let-7 axis regulates glucose metabolism. Cell.
147:81–94. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen AX, Yu KD, Fan L, Li JY, Yang C,
Huang AJ and Shao ZM: Germline genetic variants disturbing the
Let-7/LIN28 double-negative feedback loop alter breast cancer
susceptibility. PLoS Genet. 7:e10022592011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Zhang L, Fan R, Guo N, Xiong C,
Wang L, Jin S, Li W and Lu J: The polymorphism in the let-7
targeted region of the Lin28 gene is associated with increased risk
of type 2 diabetes mellitus. Mol Cell Endocrinol. 375:53–57. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnson AM and Olefsky JM: The origins and
drivers of insulin resistance. Cell. 152:673–684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Veerapur VP, Prabhakar KR, Kandadi MR,
Srinivasan KK and Unnikrishnan MK: Antidiabetic effect of Dodonaea
viscosa aerial parts in high fat diet and low dose
streptozotocin-induced type 2 diabetic rats: A mechanistic
approach. Pharm Biol. 48:1137–1148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang HC and Guarente L: SIRT1 and other
sirtuins in metabolism. Trends Endocrinol Metab. 25:138–145. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
El-Mahmoudy A, Shimizu Y, Shiina T,
Matsuyama H, El-Sayed M and Takewaki T: Successful abrogation by
thymoquinone against induction of diabetes mellitus with
streptozotocin via nitric oxide inhibitory mechanism. Int
Immunopharmacol. 5:195–207. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mondol V and Pasquinelli AE: Let's make it
happen: The role of let-7 microRNA in development. Curr Top Dev
Biol. 99:1–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thornton JE and Gregory RI: How does lin28
let-7 control development and disease? Trends Cell Biol.
22:474–482. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Frost RJ and Olson EN: Control of glucose
homeostasis and insulin sensitivity by the Let-7 family of
microRNAs. Proc Natl Acad Sci USA. 108:pp. 21075–21080. 2011;
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang C, Bao W, Rong Y, Yang H, Bowers K,
Yeung E and Kiely M: Genetic variants and the risk of gestational
diabetes mellitus: A systematic review. Hum Reprod Update.
19:376–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Petrie JR, Pearson ER and Sutherland C:
Implications of genome wide association studies for the
understanding of type 2 diabetes pathophysiology. Biochem
Pharmacol. 81:471–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rutanen J, Yaluri N, Modi S, Pihlajamäki
J, Vänttinen M, Itkonen P, Kainulainen S, Yamamoto H, Lagouge M,
Sinclair DA, et al: SIRT1 mRNA expression may be associated with
energy expenditure and insulin sensitivity. Diabetes. 59:829–835.
2010. View Article : Google Scholar : PubMed/NCBI
|