Introduction
The human embryo lung cellular protein interacting
with severe acute respiratory syndrome-coronavirus nonstructural
protein-10 (SARS-CoV nsp-10; HEPIS) gene is a novel gene that was
initially discovered by Hong et al (1) in 2008 from a cDNA library of human
embryo lung tissues. The HEPIS protein is able to interact with
SARS-CoV nsp-10 (1). SARS-CoV nsp-10
is produced by the coronavirus main protease, which cleaves
polyproteins pp1a-pp1ab during infection; this protein is able to
function as a viral transcriptase (2). The HEPIS protein consists of 147 amino
acids and has several casein kinase II phosphorylation sites
(1). In a previous study, HEPIS was
demonstrated to interact specifically with the TATA sequence of the
heat shock protein 70 promoter, suggesting that HEPIS may be
associated with gene transcriptional regulation (1). However, the expression profile and
promoter activity of HEPIS are yet to be elucidated.
Changes in the expression of specific gene products
are regulated by a wide range of mechanisms, including
transcriptional and translational regulation (3). Octamer transcription factor-1 (OCT-1),
nuclear factor κB (NF-κB) and activator protein 1 (AP-1) are
important transcription factors that serve roles in cancer cell
proliferation, survival, transformation, invasion, metastasis,
angiogenesis and chemotherapy/radiotherapy resistance (4). OCTs are a class of transcription factor
that bind to the ‘ATTTGCAT’ sequence of the gene promoter (5). OCT-1 (also termed POU2F1) is a
ubiquitously expressed transcription factor containing a POU domain
with a homeobox subdomain (6). OCT-1
serves an important regulatory role in cellular transcription via
binding to a specific promoter octamer sequence on the target genes
(7). Furthermore, OCT-1 binds to
cofactors that interact with the POU DNA-binding domain to either
positively or negatively regulate a variety of genes (8). Previous studies have reported that
OCT-1 affects the occurrence and development of several cancers,
including breast cancer (9), LNCaP
prostate cancer (10), esophageal
squamous cell carcinoma (11) and
colorectal cancer (12). NF-κB is a
dimeric transcription factor that belongs to the Rel/NF-κB family
and is formed by hetero- or homodimerization (13). NF-κB is known to serve a vital role
in the regulation of inflammation, immunity, cell proliferation and
apoptosis (13–16). AP-1, which is a dimeric
transcriptional activator composed of Jun, Fos, activating
transcription factor and musculoaponeurotic fibrosarcoma protein
subunits (17,18), serves important roles in the
regulation of cellular proliferation, transformation,
differentiation and apoptosis via binding to a common AP-1-binding
site in the target gene promoter (19,20).
In the present study, in situ RNA
hybridization and reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) were used to detect the HEPIS gene
expression profile in several organ tissues and breast cancer cell
lines. The promoter activity of the HEPIS gene was also
investigated. The first step was to identify the core HEPIS
promoter to enable subsequent determination of the important
transcription factors. The promoter region and transcription factor
binding sites of the HEPIS gene were predicted by
bioinformatics analysis. The AP-1, NF-κB and OCT-1 binding sites of
the HEPIS promoter region were identified using
site-directed mutagenesis, dual luciferase reporter assays and
chromatin immunoprecipitation (ChIP) assays, respectively.
Materials and methods
RNA in situ hybridization
A DNA microarray containing samples from 72 cases of
tumor and normal tissue was obtained from Shaanxi Chaoying
Biotechnology Co., Ltd. (cat. no. BCN721; Xian, China). The samples
were from the following 12 organs: Esophagus, stomach, colon,
rectum, liver, lung, kidney, breast, uterine cervix, ovary,
prostate and pancreas; with 3 cores positive for cancer and 3 cores
of adjacent normal tissue from each organ and one cancer tissue
core and one adjacent normal tissue core per case. The following
sense and antisense probes matching the HEPIS core
responding sequence were used: Antisense, digoxigenin (DIG)-TCT GCC
CAT ATG TCA GGA TTG GAA ATA ATG GAT −3′ and sense, DIG-ATC CAT TAT
TTC CAA TCC TGA CAT ATG GGC AGA-3′. All probes were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China). Hybridization
procedures were performed as previously described (21). Staining was scored using a 0–3+
scale. 0, no staining; 1+, 2+ and 3+ indicate increased intensity
of the staining. Sub-regions excluding necrosis, macrophages and
infiltrated neutrophils and lymphocytes were selected and scored.
The intensity score for an array spot is the mean of all its
sub-regions.
Cell culture
MDA-MB-231, MCF-7, T-47D, ZR-75-30 and 293T cells
(China Center for Type Culture Collection, Wuhan, China) were
maintained in high-glucose Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) and incubated at 37°C with 5%
CO2. 293T cells were seeded at a density of
15×104 cells/well in 6-well plates for quantitative ChIP
assays. 293T cells were seeded at a density of 5×104
cells/well in 24-well plates for luciferase assays.
RT-qPCR
Total RNA was extracted from MDA-MB-231, MCF-7,
T-47D and ZR-75-30 breast cancer cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The extracted total RNA (0.5 µg per
sample) was then used to synthesize first-strand cDNA using a
GoScript™ Reverse Transcription System kit (Promega
Corporation, Madison, WI, USA), according to the manufacturer's
protocol. The primers used for PCR were as follows: HEPIS, forward,
5′-ATGTGGCTCAGTTTGTCCTC-3′ and reverse, 5′-AGCAAGATTTCCTCCAGGTC-3′;
GAPDH, forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH was used as an internal control.
qPCR was performed using a SYBR Master Mixture (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocol using the following cycling conditions:
95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C
for 30 sec. The expression of HEPIS was analyzed as previously
described (22).
Plasmid construction
The promoter sequence of the HEPIS gene
(pHEPIS) was obtained by PCR from MCF-7 cell genomic DNA using the
following primers: pHEPIS-F1.7k (−1334), forward 5′-ATCCTCGAGCATCACAAGTAGGGCAGCAT-3′;
pHEPIS-F1.6k (−1060), forward 5′-ATCCTCGAGGAGTCTTCAAAGGGAGTG-3′;
pHEPIS-F1.4k (−1203), forward 5′-ATCCTCGAGTCCTGGTATGCCAAGAAA-3′;
pHEPIS-F1.3k (−899), forward 5′-ATCCTCGAGCAAGCTGATAGCCACCAA-3′;
pHEPIS-F1.1k (−759), forward 5′-ATCCTCGAGAGGTTGGCAGGCCGGATAT-3′;
pHEPIS-F0.6k (−279), forward 5′-ATCCTCGAGCGAAGAGGAGGGAGGTAG-3′;
pHEPIS-R (+373), reverse 5′-AGTAAGCTTACTTCGCACCTTCGGCTA-3′.
PCR was performed using pyrobest DNA polymerase (Takara
Biotechnology Co., Ltd.). The PCR amplification reaction system
conditions and PCR products were purified as previously described
(23). Purified products were cloned
into the XhoI (CTCGAG) and HindIII
(AAGCTT)
restriction enzyme sites of the pGL3-basic vector (Promega
Corporation) using T4 DNA ligase (Sangon Biotech Co., Ltd.)
according to the manufacturer's protocol.
In the present study, the transcription factor
database TRANSFAC (www.cbrc.jp/research/db/TFSEARCH.html) was used for
the search, and several AP-1, NF-κB and OCT-1 transcription
factor-binding sites were predicted within the HEPIS
promoter region. Site-directed mutageneses of the OCT-1
(−1236/−1223, negative numbers indicate that it is upstream of the
transcription initiation site), NF-κB (−1186/−1176) and C-JUN
(−856/−846) binding sites in the HEPIS promoter were performed
using a Quick Change Site-Directed Mutagenesis kit (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA) according to
manufacturer's protocol, using the following primers:
pHEPIS-OCT-1-M, forward
5′-TTATAGGTGTCAAATTCATCATCACCATCAAAACTGCGTGCTTCTGCACTGAAACA-3′ and
reverse 5′-TGTTTCAGTGCAGAAGCACGCAGTTTTGATGGTGATGATGAATTTGACACCTATAA-3′;
pHEPIS-NF-κB-M, forward 5′-GAGTCTTCAAAGGGAGTGGAATTACCTGGATCTTCTGTTG-3′ and
reverse 5′-CAACAGAAGATCCAGGTAATTCCAACTCCCTTTGAAGACTC-3′;
pHEPIS-C-JUN-M, forward 5′-AATAACAAATTCATCATTGTTAGTTTGTAGCAGGATTGCACTGGAGACAGAGATTCC-3′
and reverse 5′-GGAATCTCTGTCTCCAGTGCAATCCTGCTACAAACTAACAATGATGAATTTGTTATT-3′.
Underlined base pairs indicate mutation sites.
Transfection and dual luciferase
reporter assay
293T cells were cotransfected with 1 µg pGL3-basic
vector, pHEPIS-1.7K, pHEPIS-1.6K, pHEPIS-1.4K, pHEPIS-1.3K,
pHEPIS-1.1K, pHEPIS-0.6K, pHEPIS-1.7K-M-OCT-1, pHEPIS-1.6K-M-NF-κB,
pHEPIS-1.3k-M-C-JUN or pHEPIS-1.7K-3M and 0.2 µg pRL-TK (Promega
Corporation) plasmid DNA/well in 24-well plates using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The luciferase activity of the extracts
was assessed 24 h following transfection using a Betascope analyzer
Infinite M200, (Tecan Group Ltd., Männedorf, Switzerland) and
analyzed as previously described (23). The pRL-TK plasmid containing the
Renilla luciferase gene was used as an internal control.
ChIP assays
ChIP assays were performed according to the
manufacturer's protocol using a Millipore ChIP assay kit (EMD
Millipore, Billerica, MA, USA). The following primary antibodies
were used: Rabbit polyclonal antibodies against NF-κB p65 (cat. no.
ab7970, 1:200), OCT-1 (cat. no. ab66132, 1:200; both Abcam,
Cambridge, UK) and C-JUN (cat. no. sc-1694, 1:100), and anti-rabbit
normal immunoglobulin G (cat. no. sc-2345, 1:100; both Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was used as an negative
control. The above antibodies were used per chromatin sample and
rotated overnight at 4°C. Protein A/G Agarose/Salmon Sperm DNA
Secondary antibody (1:400; cat. nos. 16-157 and 16-201; EMD
Millipore) was added per sample for 1 h at 4°C with rotation. The
amount of each specific DNA fragment in the immunoprecipitates was
determined using PCR reactions with the following primers: OCT-1,
forward 5′-ATGTAATCCAGTAGCCTGTC-3′ and reverse
5′-CTCCCTTTGAAGACTCTGA-3′; NF-κB, forward
5′-TTCAGAGTCTTCAAAGGGAG-3′ and reverse 5′-GCATACCAGGAGACAATAAAC-3′;
C-JUN, forward 5′-GCCACCAACAATAACAAA-3′ and reverse
5′-AGGAGGACATTCACTTGC-3′. The PCR was performed using a PCR Master
Mix (Sangon Biotech, Shanghai, China) according to the
manufacturer's protocol using the following cycling conditions:
95°C for 30 sec, followed by 30 cycles of 95°C for 30 sec, 60°C for
30 sec and 72°C for 10 sec; 72°C for 5 min.
Statistical analysis
Statistical analysis was performed using SPSS 9.0
software (SPSS, Inc., Chicago, IL, USA). Data are presented as the
mean ± standard deviation. Student's t-test and one-way analysis of
variance followed by a Dunnett's test were used to analyze data.
P<0.05 was considered to indicate a statistically significant
difference.
Results
HEPIS expression profile in tissues
and breast cancer cells
The HEPIS expression profile was detected by
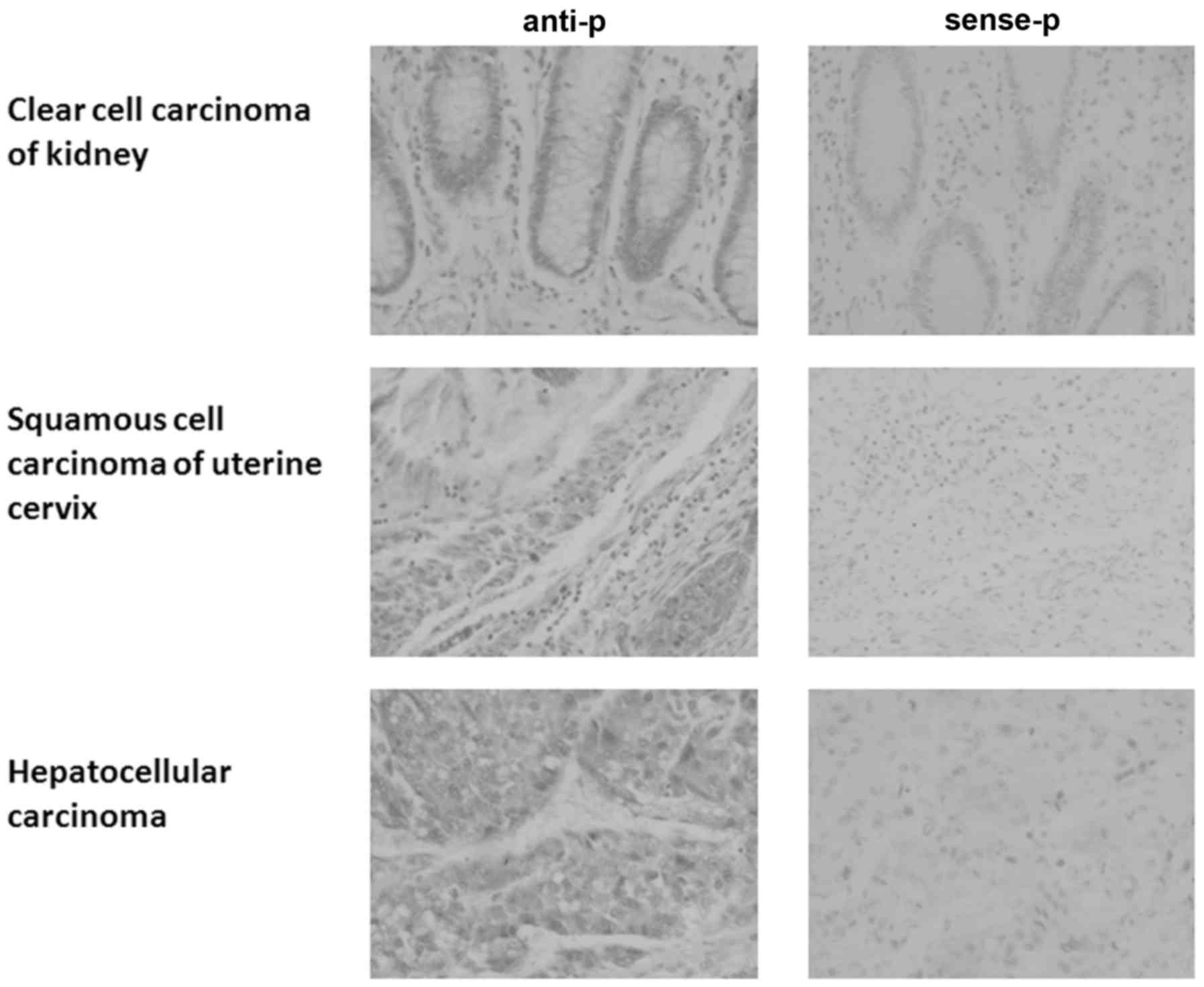
RNA in situ hybridization in a tissue microarray.
HEPIS expression in esophageal squamous cell carcinoma and
rectal adenocarcinoma tissues was the opposite of that in normal
esophageal and rectal tissues (Table
I; Fig. 1); HEPIS
expression was positive in esophageal squamous cell carcinoma and
negative in normal esophageal tissue, whereas it was positive in
normal rectal tissue and negative in rectal adenocarcinoma.
HEPIS was positively expressed in tumor and normal tissues
from the stomach, liver, colon, prostate, lung, uterine cervix and
pancreas (Table I). The expression
of HEPIS was positive in some tumor and normal tissues of
the kidneys and ovaries and negative in others. HEPIS was
positively expressed in nonspecific infiltrating duct carcinoma of
the breast and partial positive expression was observed in normal
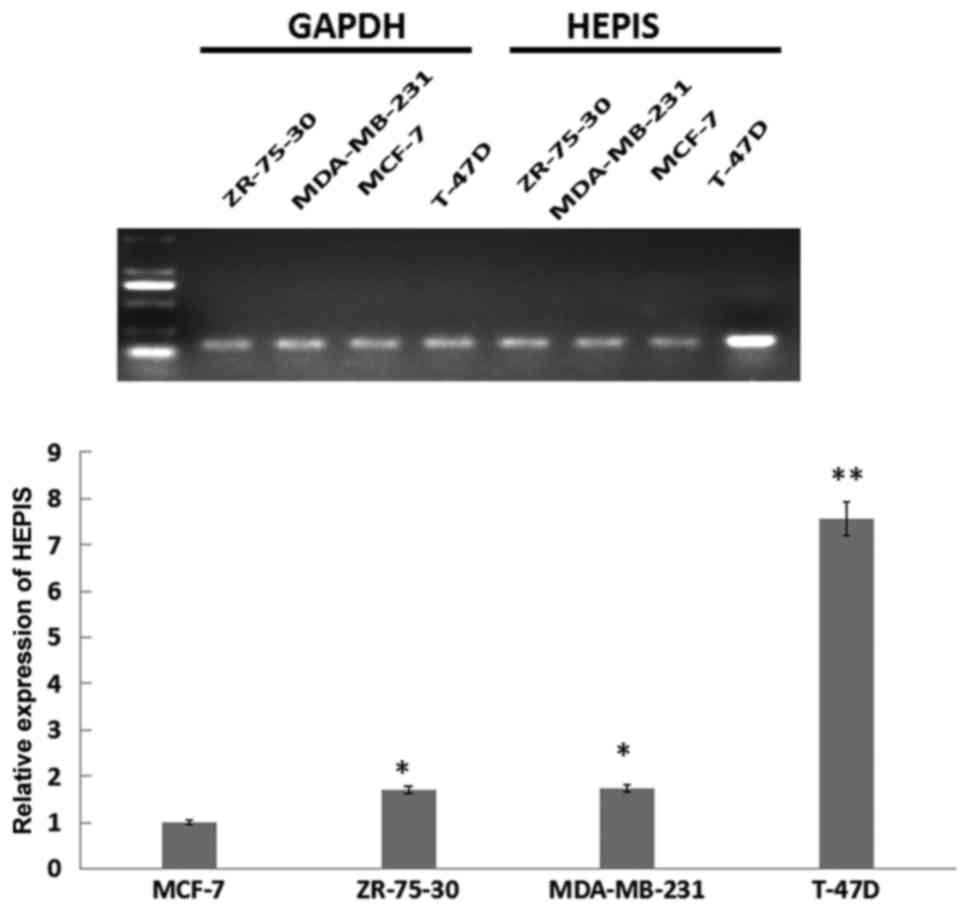
breast tissue. HEPIS expression levels in four human breast
cancer cell lines was examined using RT-qPCR (Fig. 2). The expression of HEPIS was
significantly increased in the osteolytic breast cancer T-47D cell
line compared with ZR-75-30, MDA-MB-231 and MCF-7 cells.
HEPIS mRNA levels in T-47D cells were ~8-fold higher
compared with MCF-7 cells (P<0.01), and in ZR-75-30, MDA-MB-231
cells were ~1.8-fold higher compared with MCF-7 cells (P<0.05).
These results suggest that HEPIS is expressed at different levels
in various organs and breast cancer cell lines.
 | Table I.HEPIS expression in multiple organ
cancer and normal tissue. |
Table I.
HEPIS expression in multiple organ
cancer and normal tissue.
| Organ | Pathology
diagnosis | Tissues/samples
(n) | HEPIS mRNA-positive
tumors, n (+/++/+++) | HEPIS mRNA-negative
tumors, n |
|---|
| Esophagus | Squamous cell
carcinoma | 3 | 3 (0/3/0) | 0 |
|
| Normal tissue | 3 | 0 | 3 |
| Stomach | Adenocarcinoma | 3 | 3 (1/2/0) | 0 |
|
| Normal tissue | 3 | 3 (0/1/2) | 0 |
| Colon | Adenocarcinoma | 3 | 3 (0/2/1) | 0 |
|
| Normal tissue | 3 | 3 (0/0/3) | 0 |
| Rectum | Adenocarcinoma | 3 | 0 | 3 |
|
| Normal tissue | 3 | 3 (0/3/0) | 0 |
| Liver | Hepatocellular
carcinoma | 3 | 3 (0/2/1) | 0 |
|
| Normal tissue | 3 | 3 (0/0/3) | 0 |
| Lung | Squamous cell
carcinoma | 3 | 3 (0/3/0) | 0 |
|
| Normal tissue | 3 | 3 (3/0/0) | 0 |
| Kidney | Clear cell
carcinoma | 3 | 2 (2/0/0) | 1 |
|
| Normal tissue | 3 | 1 (1/0/0) | 2 |
| Breast | Non-specific
infiltrating duct carcinoma | 3 | 3 (2/1/0) | 0 |
|
| Normal tissue | 3 | 2 (1/1/0) | 1 |
| Uterine cervix | Squamous cell
carcinoma | 3 | 3 (3/0/0) | 0 |
|
| Normal tissue | 3 | 3 (3/0/0) | 0 |
| Ovary | Serous
cystadenocarcinoma | 3 | 1 (1/0/0) | 2 |
|
| Normal tissue | 3 | 1 (1/0/0) | 2 |
| Prostate | Adenocarcinoma | 3 | 3 (1/1/1) | 0 |
|
| Normal tissue | 3 | 3 (2/1/0) | 0 |
| Pancreas | Duct
adenocarcinoma | 3 | 3 (3/0/0) | 0 |
|
| Normal tissue | 3 | 3 (0/3/0) | 0 |
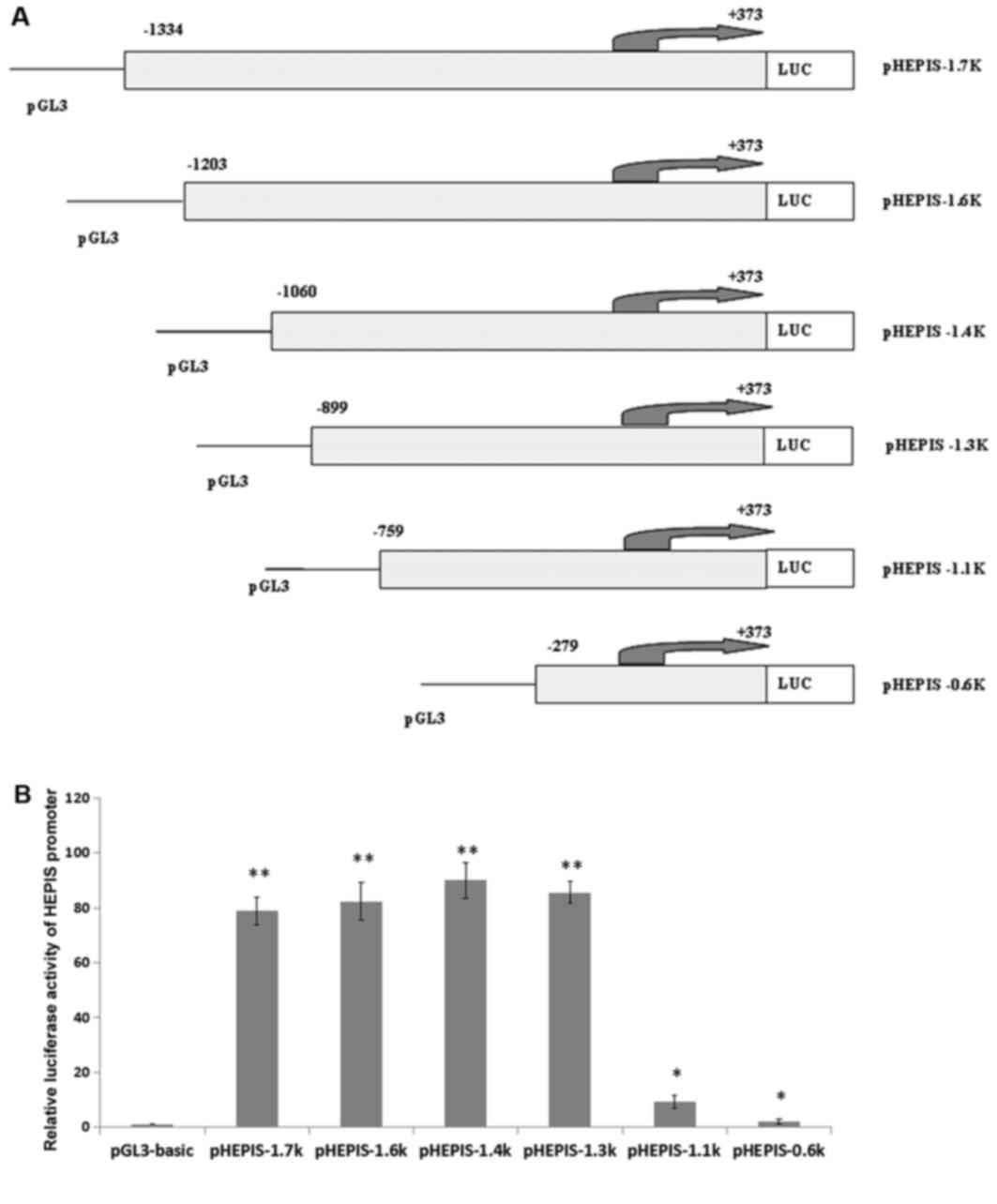
Cloning and activity of the human
HEPIS promoter
To understand the mechanism by which HEPIS
gene transcripts are expressed, dual luciferase reporter assays
were used to detect HEPIS promoter activity. A total of six
different truncated lengths of the HEPIS promoter regulatory
sequences were amplified and the PCR products were cloned into the
pGL3-basic vector (Fig. 3A). Dual
luciferase reporter assay analysis of the six recombined plasmids
revealed that the −1334/+373, −1203/+373, −1060/+373, and −899/+373
bp reporter gene fragments exhibited higher activity levels
compared with pGL3-basic (P<0.01); and the −759/+373 bp and
−279/+373 bp reporter gene fragments exhibited higher activity
levels compared with pGL3-basic (P<0.05; Fig. 3B).
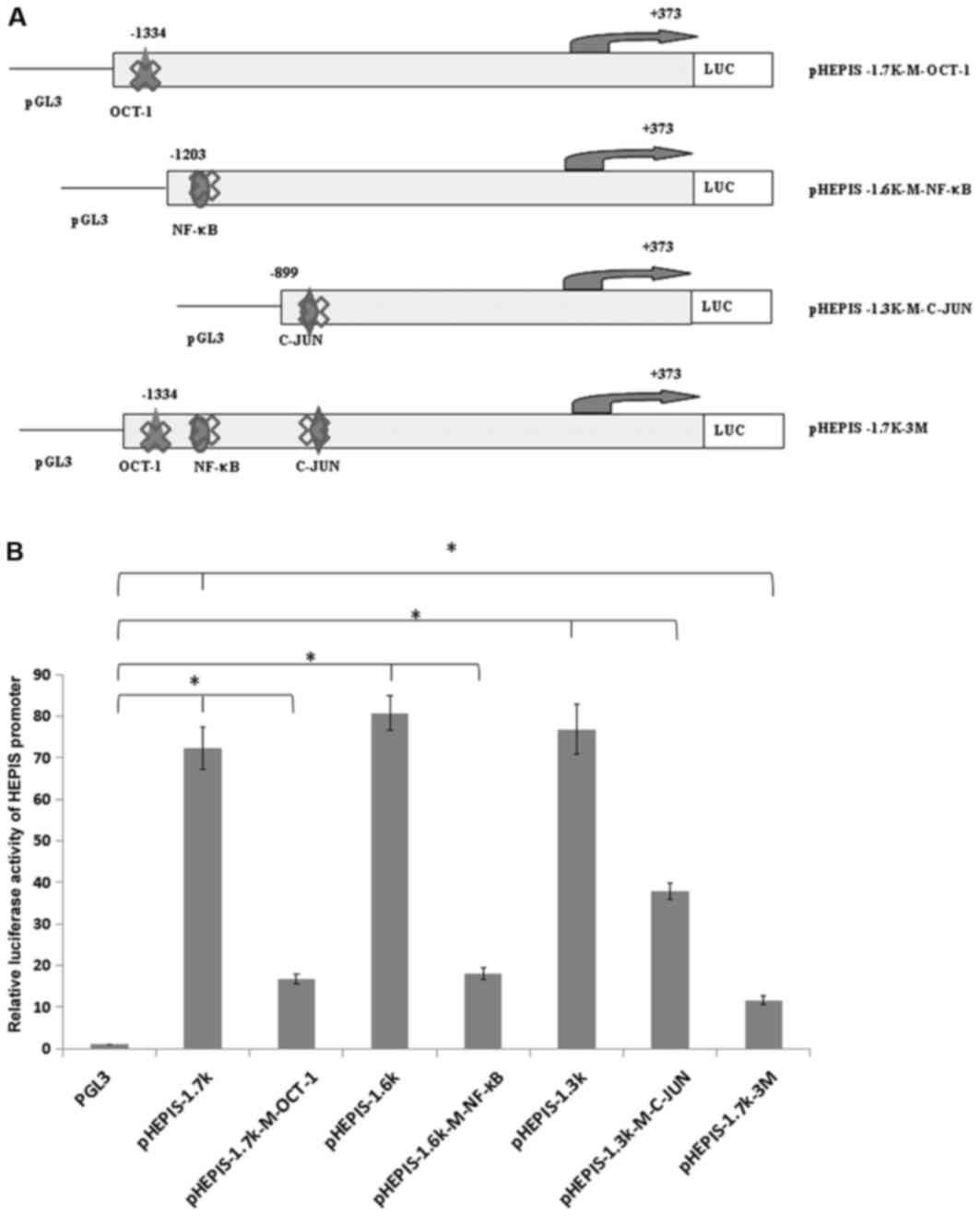
Mutations at transcription factor
binding sites and luciferase activity analysis
To investigate whether these putative response
elements regulate the transcription of HEPIS, the OCT-1
(5′-CTATTTGCTTCTG-3′, −1236/−1223 bp), NF-κB (5′-GGAATCCCCT-3′,
−1186/−1176bp), and C-JUN (5′-TTGAGTCAGG-3′, −856/−846bp) response
elements on the human HEPIS promoter were mutated to generate
pHEPIS-1.7K-M-OCT-1, pHEPIS-1.6K-M-NF-κB and pHEPIS-1.3K-M-C-JUN,
which were constructed individually (Fig. 4A). The dual luciferase assay results
demonstrated that the luciferase activities of pHEPIS-1.7K-M-OCT-1,
pHEPIS-1.6K-M-NF-κB and pHEPIS-1.3k-M-C-JUN were significantly
decreased compared with the activities of pHEPIS-1.7K, pHEPIS-1.6K
and pHEPIS-1.3K, respectively (P<0.05; Fig. 4B), suggesting that C-JUN, OCT-1 and
NF-κB activate the reporter. Furthermore, the OCT-1, NF-κB and
C-JUN binding elements of the HEPIS promoter were
simultaneously mutated to generate pHEPIS-1.7K-3M. When all three
sites were mutated, the pHEPIS-1.7K-3M promoter activity was
significantly decreased compared with the pHEPIS-1.7K (P<0.05;
Fig. 4B); however, the level of
suppression with the three mutations did not exceed the combined
level of suppression by the individual point mutations, which
suggests that the three mutations act jointly. Taken together,
these results suggest that the OCT-1, NF-κB and C-JUN sites serve
an important role in inhibiting the transcriptional activity of
HEPIS.
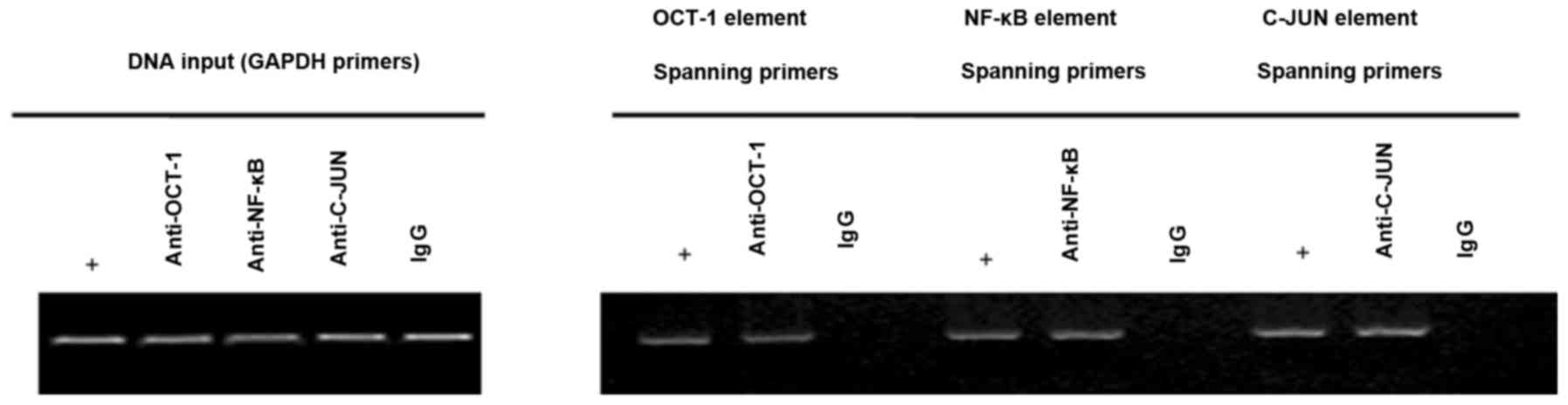
Identification of transcription
factors in the HEPIS promoter
Identifying the transcription factor binding sites
within the HEPIS promoter region is important for
determining the mechanism of HEPIS gene transcription. To
determine whether OCT-1, NF-κB and C-JUN were able to bind to the
endogenous HEPIS promoter, a ChIP assay was performed to
investigate transcription factor binding. The results indicated
that OCT-1, NF-κB and C-JUN bind to the endogenous HEPIS
promoter in 293T cells, which suggests that they may serve an
important role in regulating HEPIS expression (Fig. 5).
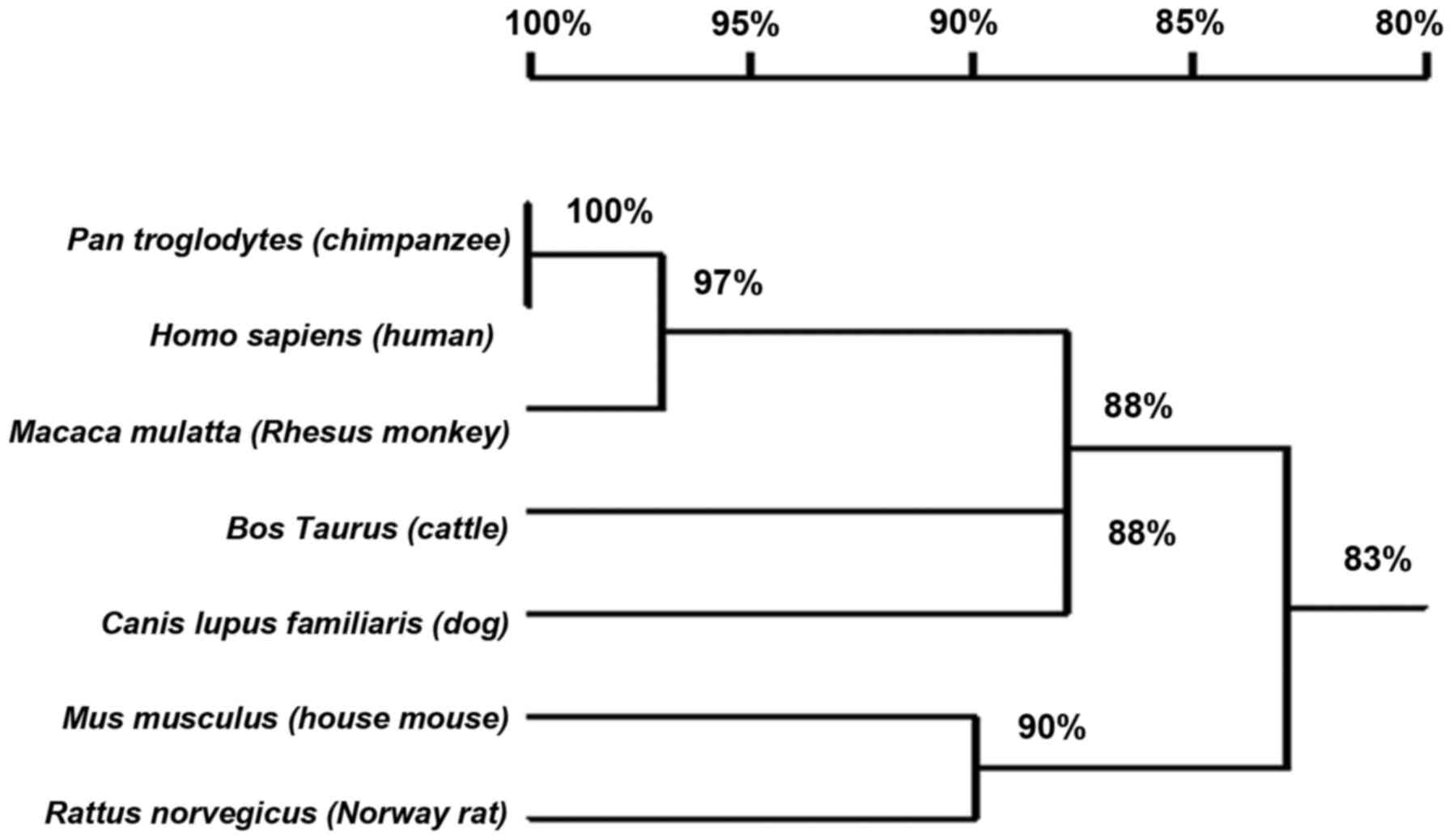
Analysis of HEPIS promoters in
multiple species
Table II lists the
putative HEPIS promoter among various species with the same
sequence length. The HEPIS promoter is conserved among
vertebrates (Fig. 6). The sequence
of the Homo sapiens (human) HEPIS promoter shares the
highest homology (100%) with that of Pan troglodytes
(chimpanzees). The HEPIS promoters of Rattus
norvegicus (Norway rats), Mus musculus (house mice),
Bos taurus (cattle), chimpanzees, humans, Canis lupus
familiaris (dogs) and Macaca mulatta (Rhesus monkeys)
were analyzed using TRANSFAC and several AP-1, C-JUN, C-Fos, NF-κB
and OCT-1 transcription factor binding sites were predicted within
the promoter region (Table II).
 | Table II.Analysis of HEPIS promoters in
multiple species. |
Table II.
Analysis of HEPIS promoters in
multiple species.
|
|
| Transcriptional
factor binding sites, n |
|---|
|
|
|
|
|---|
| Species | Context of
HEPIS promoter (−2.0k) | OCT-1 | NF-κB | AP-1 | C-JUN | C-Fos |
|---|
| Rattus
norvegicus (Norway rat) | Chr3:
91195981-91197981 [-] | 11 | 0 | 6 | 2 | 0 |
| Mus musculus
(house mouse) | Chr2:
101629105–101631105 [-] | 13 | 11 | 6 | 2 | 1 |
| Homo Sapiens
(human) | Chr11:
36592229–36594229 [+] | 14 | 5 | 5 | 2 | 0 |
| Bos Taurus
(cattle) | Chr15:
67842229–67844229 [+] | 17 | 3 | 9 | 4 | 1 |
| Pan
troglodytes (chimpanzee) | Chr11:
36583771–36585771 [+] | 14 | 6 | 6 | 2 | 0 |
| Canis lupus
familiaris (dog) | Chr18:
31618122–31620122 [-] | 15 | 3 | 7 | 4 | 3 |
| Macaca
mulatta (rhesus monkey) | Chr14:
29326253–29328253 [-] | 14 | 4 | 5 | 2 | 0 |
Discussion
It has previously been reported that HEPIS is
able to inhibit the proliferation of HeLa cells and may serve as an
anti-oncoprotein (1). HEPIS
is also able to inhibit the expression of the chloramphenicol
acetyltransferase gene and may function as a factor of
transcriptional repression (1). The
aim of the present study was to determine the expression profile of
the HEPIS gene and further elucidate the mechanism by which
HEPIS transcriptional levels differ. RNA in situ
hybridization (RISH) is a method of identifying the mRNA
transcriptional expression pattern within the cytoplasm by
hybridizing the sequence of interest to a labeled probe (24). Probes include radioactive probes and
non-radioactive probes and RISH experiments performed with
non-radioactive probes have several advantages over the radioactive
procedures, including signal resolution, safety, shelf-life and
cost (25). Due to the limited
availability of the HEPIS antibody, the expression of the
HEPIS gene in cancer and adjacent normal tissues in 12
organs was assessed using RNA in situ hybridization with a
specific digoxigenin-labelled probe. However, due to the limited
number of specimens available, it was necessary to further increase
the number of specimens analyzed in order to obtain accurate
results. HEPIS expression levels in four human breast cancer cell
lines was examined using RT-qPCR, however, HEPIS expression in
other types of cell lines remains unknown. Determining the
differential expression of HEPIS allows analysis of its function in
a variety of diseases.
Promoters control gene transcription. They may be
located upstream of the gene transcription start site and can be
very long (26). The binding of
transcription factors to a promoter is an important mechanism by
which gene expression is controlled (26). Investigating HEPIS promoter
activity revealed that the luciferase activity varied between
pHEPIS-1.3k and pHEPIS-1.1k, suggesting that transcriptional
regulation occurs at the-899/−759 bp region of the promoter.
Furthermore, the results suggest that mutations of C-JUN
(TTGAGTCAGG, −856/−846 bp), OCT-1 (CTATTTGCTTCTG, −1236/−1223 bp)
and NF-κB (GGAATCCCCT, −1186/−1176 bp) result in a marked reduction
in luciferase activity, which indicated that C-JUN, OCT-1 and NF-κB
are activators. However, no significant changes in luciferase
activity were observed following truncation of the −1334/1203 bp
and −1203/−1060 bp regions. These results suggest that the
−1334/1203 bp and −1203/−1060 bp regions also contain
repressor-binding sites. The findings of the present study indicate
that the apparent changes in transcriptional activity of the
HEPIS gene may result from complex interactions of different
transcription factors with the promoter. The association of
HEPIS gene and the above transcription factors maybe
widespread and therefore, further study is required.
In the present study, sequence analysis identified
numerous transcription factor-binding sites within the HEPIS
promoter sequence. Of these, OCT-1 NF-κB and C-JUN were
ubiquitously expressed; these have previously been reported to
serve a variety of roles in the progression of numerous cancers
(5,9,16,17,19,20).
The results of the ChIP assay indicated that OCT-1, NF-κB and C-JUN
are able to bind to the endogenous HEPIS promoter in 293T cells.
Several AP-1, C-JUN, C-Fos, NF-κB and OCT-1 transcription
factor-binding sites were predicted within the putative
HEPIS promoter in various species.
In conclusion, the results of this present research
revealed that HEPIS has different expression levels in multiple
types of cancer and normal tissues, and four breast cancer cell
lines; and the OCT-1, NF-κB and C-JUN transcription factors are
associated with transcriptional regulation of the HEPIS
gene. These findings provide further insight into the expression
profile and the mechanism of HEPIS gene transcriptional
regulation.
Acknowledgements
The present study was supported by the General
Higher Education Young Talents Program of Hebei Province (grant no.
BJ2014027) and the National Natural Science Foundation of China
(grant no. 81302323).
References
|
1
|
Hong M, Li W, Wang L, Jiang L, Liu L, Zhao
H and Li Q: Identification of a novel transcriptional repressor
(HEPIS) that interacts with nsp-10 of SARS coronavirus. Viral
immunol. 21:153–162. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sawicki SG, Sawicki DL, Younker D, Meyer
Y, Thiel V, Stokes H and Siddell SG: Functional and genetic
analysis of coronavirus replicase-transcriptase proteins. PLoS
Pathog. 1:e392005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Edwards R, Machina A, McGregor G and van
den Driessche P: A modelling framework for gene regulatory networks
including transcription and translation. Bull Math Biol.
77:953–983. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shanmugam MK, Nguyen AH, Kumar AP, Tan BK
and Sethi G: Targeted inhibition of tumor proliferation, survival,
and metastasis by pentacyclic triterpenoids: Potential role in
prevention and therapy of cancer. Cancer Lett. 320:158–170. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sterling K and Bresnick E: Oct-1
transcription factor is a negative regulator of rat CYP1A1
expression via an octamer sequence in its negative regulatory
element. Mol Pharmacol. 49:329–337. 1996.PubMed/NCBI
|
|
6
|
Lee MC, Toh LL, Yaw LP and Luo Y:
Drosophila octamer elements and Pdm-1 dictate the coordinated
transcription of core histone genes. J Biol Chem. 285:9041–9053.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang P, Wang Q, Sun J, Wu J, Li H, Zhang
N, Huang Y, Su B, Li RK, Liu L, et al: POU homeodomain protein
Oct-1 functions as a sensor for cyclic AMP. J Biol Chem.
284:26456–26465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malin S, Linderson Y, Almqvist J, Ernberg
I, Tallone T and Pettersson S: DNA-dependent conversion of Oct-1
and Oct-2 into transcriptional repressors by Groucho/TLE. Nucleic
Acids Res. 33:4618–4625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou C, Tong Y, Wawrowsky K, Bannykh S,
Donangelo I and Melmed S: Oct-1 induces pituitary tumor
transforming gene expression in endocrine tumors. Endocr-Relat
Cancer. 15:817–831. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Obinata D, Takayama K, Urano T, Murata T,
Kumagai J, Fujimura T, Ikeda K, Horie-Inoue K, Homma Y, Ouchi Y, et
al: Oct1 regulates cell growth of LNCaP cells and is a prognostic
factor for prostate cancer. Int J Cancer. 130:1021–1028. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Zhu S, Shen M, Liu J, Wang M, Li
C, Wang Y, Deng A and Mei Q: STAT3 is involved in esophageal
carcinogenesis through regulation of Oct-1. Carcinogenesis.
34:678–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang YP, Song GH, Chen J, Xiao C, Li C,
Zhong L, Sun X, Wang ZW, Deng GL, Yu FD, et al: Elevated OCT1
participates in colon tumorigenesis and independently predicts poor
prognoses of colorectal cancer patients. Tumour Biol. 37:3247–3255.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karin M and Ben-Neriah Y: Phosphorylation
meets ubiquitination: The control of NF-[kappa] B activity. Annu
Rev Immunol. 18:621–663. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gebel HM, Braun DP, Tambur A, Frame D,
Rana N and Dmowski WP: Spontaneous apoptosis of endometrial tissue
is impaired in women with endometriosis. Fertil Steril.
69:1042–1047. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huxford T, Malek S and Ghosh G: Structure
and mechanism in NF-kappa B/I kappa B signaling. Cold Spring Harb
Symp Quant Biol. 64:533–540. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldwin AS Jr: The NF-kappa B and I kappa
B proteins: New discoveries and insights. Annu Rev Immunol.
14:649–683. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shaulian E and Karin M: AP-1 in cell
proliferation and survival. Oncogene. 20:2390–2400. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eferl R and Wagner EF: AP-1: A
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li GC, Gustafson-Brown C, Hanks SK, Nason
K, Arbeit JM, Pogliano K, Wisdom RM and Johnson RS: c-Jun is
essential for organization of the epidermal leading edge. Dev Cell.
4:865–877. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen G, Jeong WS, Hu R and Kong AN:
Regulation of Nrf2, NF-kappaB, and AP-1 signaling pathways by
chemopreventive agents. Antioxid Redox Signal. 7:1648–1663. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu F, Yang S, Lv S, Peng Y, Meng L, Gou L
and Zhang X: Analysis of AC3-33 gene expression in multiple organ
cancer and adjacent normal tissue by RNA in situ hybridization.
Oncol Lett. 9:2795–2798. 2015.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu F, Meng Y, Gou L and Zhang X: Analysis
of promoters and CREB/AP-1 binding sites of the human TMEM174 gene.
Exp Ther Med. 6:1290–1294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Denijn M, Schuurman HJ, Jacobse KC and De
Weger RA: In situ hybridization: A valuable tool in diagnostic
pathology. APMIS. 100:669–681. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vale G and Dell'Orto P: Non-radioactive
nucleic acid probes: Labeling and detection procedures. Liver.
12:243–251. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Farnham PJ: Insights from genomic
profiling of transcription factors. Nat Rev Genet. 10:605–616.
2009. View
Article : Google Scholar : PubMed/NCBI
|