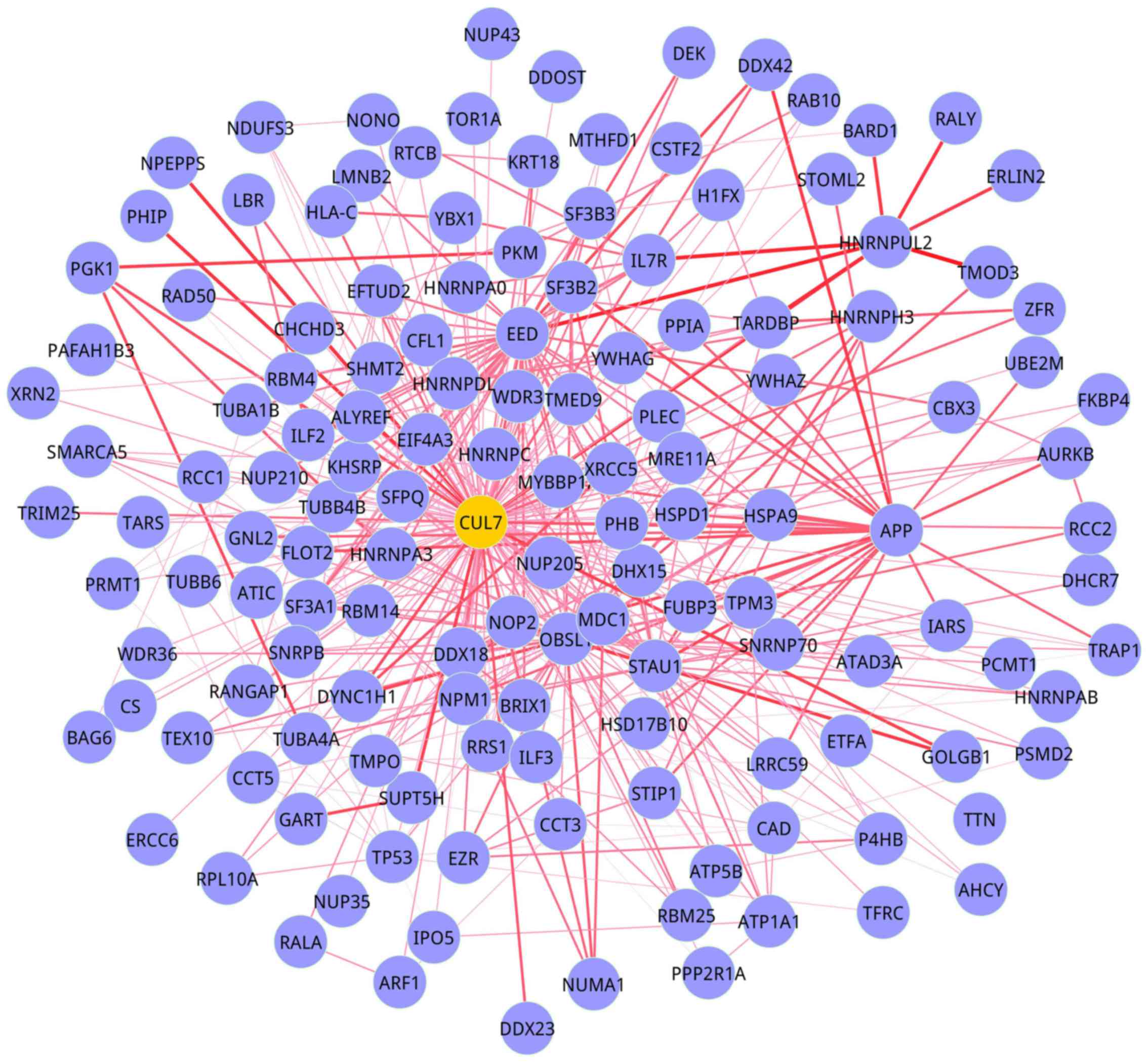
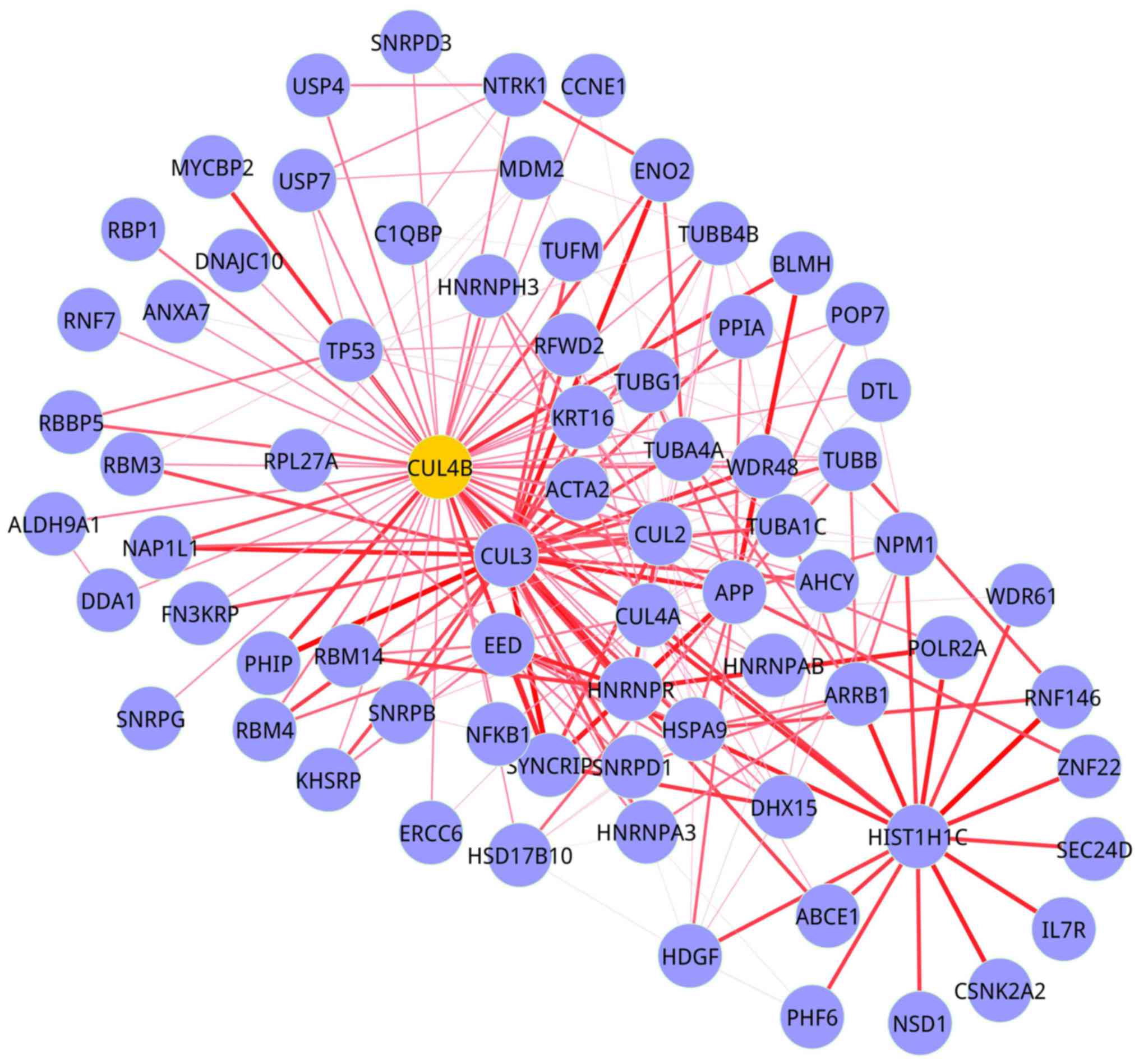
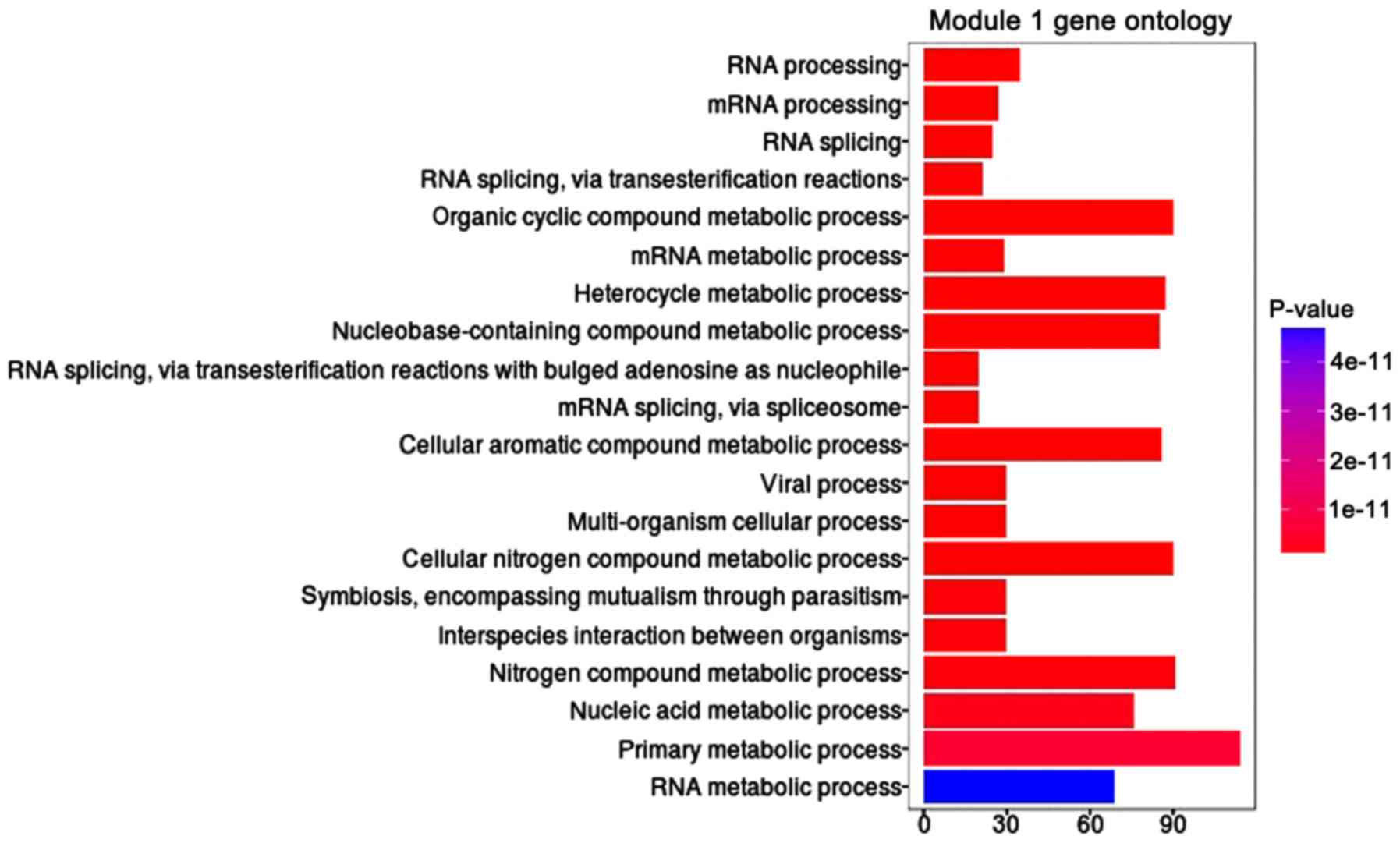
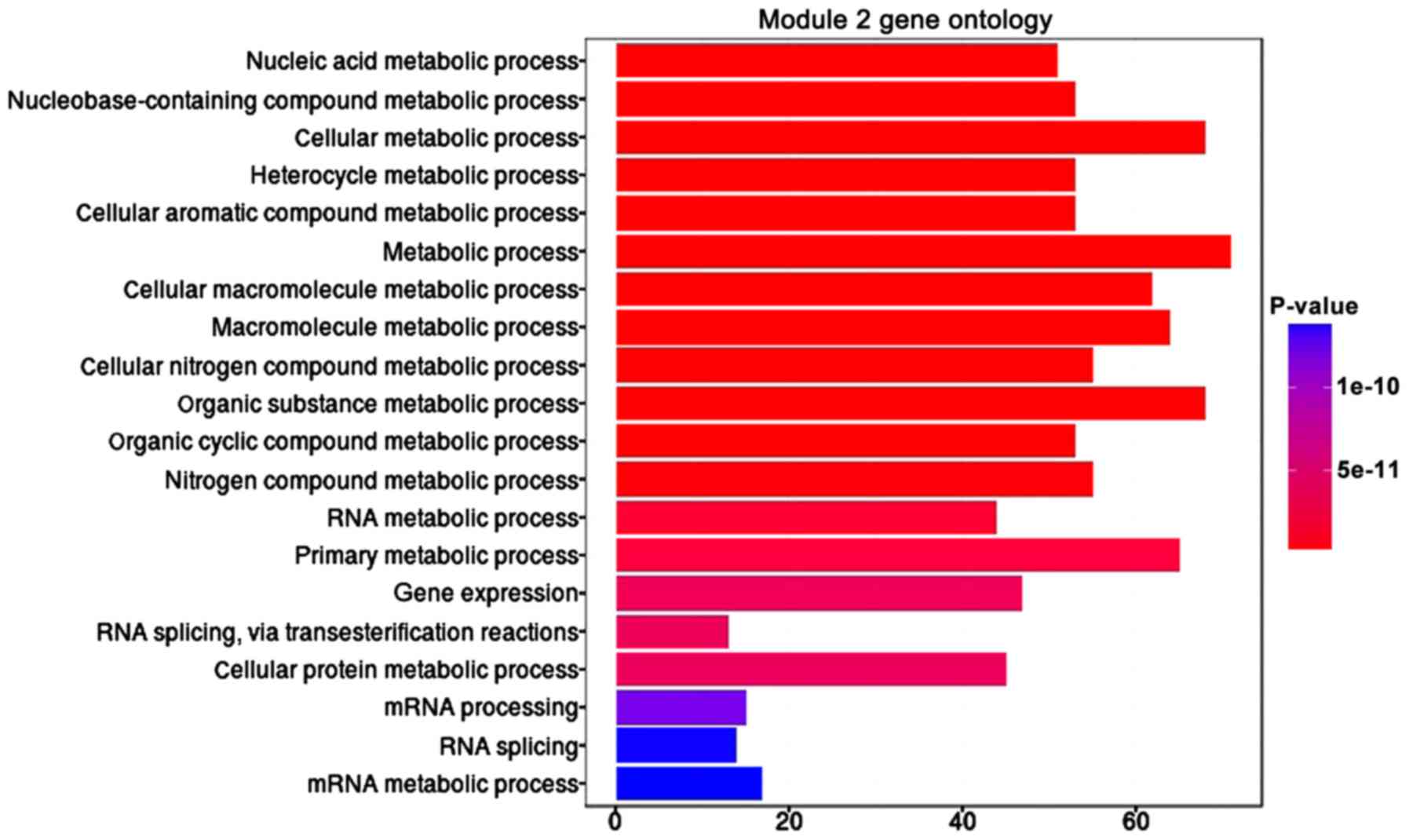
|
1
|
Ruygrok PN and Serruys PW: Intracoronary
stenting. From concept to custom. Circulation. 94:882–890. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bønaa KH, Mannsverk J, Wiseth R, Aaberge
L, Myreng Y, Nygård O, Nilsen DW, Kløw NE, Uchto M, Trovik T, et al
NORSTENT Investigators, : Drug-eluting or bare-metal stents for
coronary artery disease. N Engl J Med. 375:1242–1252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Serruys PW, Kutryk MJ and Ong AT:
Coronary-artery stents. N Engl J Med. 354:483–495. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moses JW, Leon MB, Popma JJ, Fitzgerald
PJ, Holmes DR, O'Shaughnessy C, Caputo RP, Kereiakes DJ, Williams
DO, Teirstein PS, et al SIRIUS Investigators, : Sirolimus-eluting
stents versus standard stents in patients with stenosis in a native
coronary artery. N Engl J Med. 349:1315–1323. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Laarman GJ, Suttorp MJ, Dirksen MT, van
Heerebeek L, Kiemeneij F, Slagboom T, van der Wieken LR, Tijssen
JG, Rensing BJ and Patterson M: Paclitaxel-eluting versus uncoated
stents in primary percutaneous coronary intervention. N Engl J Med.
355:1105–1113. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Authors/Task Force M, ; Windecker S, Kolh
P, Alfonso F, Collet JP, Cremer J, Falk V, Filippatos G, Hamm C,
Head SJ, et al: 2014 ESC/EACTS Guidelines on myocardial
revascularization: The Task Force on Myocardial Revascularization
of the European Society of Cardiology (ESC) and the European
Association for Cardio-Thoracic Surgery (EACTS)Developed with the
special contribution of the European Association of Percutaneous
Cardiovascular Interventions (EAPCI). Eur Heart J. 35:2541–2619.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huan T, Zhang B, Wang Z, Joehanes R, Zhu
J, Johnson AD, Ying S, Munson PJ, Raghavachari N, Wang R, et al
Coronary artery disease genome wide replication and meta-analysis
(CARDIoGRAM) consortium, International Consortium for Blood
Pressure GWAS (ICBP), : A systems biology framework identifies
molecular underpinnings of coronary heart disease. Arterioscler
Thromb Vasc Biol. 33:1427–1434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gargalovic PS, Imura M, Zhang B, Gharavi
NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, et al:
Identification of inflammatory gene modules based on variations of
human endothelial cell responses to oxidized lipids. Proc Natl Acad
Sci USA. 103:pp. 12741–12746. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akavia UD and Benayahu D: Meta-analysis
and profiling of cardiac expression modules. Physiol Genomics.
35:305–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Segal E, Shapira M, Regev A, Pe'er D,
Botstein D, Koller D and Friedman N: Module networks: Identifying
regulatory modules and their condition-specific regulators from
gene expression data. Nat Genet. 34:166–176. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanay A and Shamir R: Computational
expansion of genetic networks. Bioinformatics. 17 Suppl
1:S270–S278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pe'er D, Regev A and Tanay A: Minreg:
Inferring an active regulator set. Bioinformatics. 18 Suppl
1:S258–S267. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma X, Gao L, Karamanlidis G, Gao P, Lee
CF, Garcia-Menendez L, Tian R and Tan K: Revealing pathway dynamics
in heart diseases by analyzing multiple differential networks. PLOS
Comput Biol. 11:e10043322015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nayak RR, Kearns M, Spielman RS and Cheung
VG: Coexpression network based on natural variation in human gene
expression reveals gene interactions and functions. Genome Res.
19:1953–1962. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vizeacoumar FJ, van Dyk N, Vizeacoumar F
S, Cheung V, Li J, Sydorskyy Y, Case N, Li Z, Datti A, Nislow C, et
al: Integrating high-throughput genetic interaction mapping and
high-content screening to explore yeast spindle morphogenesis. J
Cell Biol. 188:69–81. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao Y, Xu C, Guan J, Ping Y, Fan H, Li Y,
Zhao H and Li X: Discovering dysfunction of multiple microRNAs
cooperation in disease by a conserved microRNA co-expression
network. PLoS One. 7:e322012012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Robinson LN and Towle HC: ChREBP*Mlx
is the principal mediator of glucose-induced gene expression in the
liver. J Biol Chem. 281:28721–28730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rifai N and Ridker PM: Proposed
cardiovascular risk assessment algorithm using high-sensitivity
C-reactive protein and lipid screening. Clin Chem. 47:28–30.
2001.PubMed/NCBI
|
|
19
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix genechip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al The Gene Ontology Consortium, : Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stettler C, Wandel S, Allemann S, Kastrati
A, Morice MC, Schömig A, Pfisterer ME, Stone GW, Leon MB, de Lezo
JS, et al: Outcomes associated with drug-eluting and bare-metal
stents: A collaborative network meta-analysis. Lancet. 370:937–948.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miki K, Fujii K, Kawasaki D, Fukunaga M,
Nishimura M, Horimatsu T, Saita T, Tamaru H, Imanaka T, Shibuya M,
et al: Effect of bare-metal nitinol stent implantation and
paclitaxel-eluting nitinol stent implantation on vascular response
in the superficial femoral artery lesion assessed on intravascular
ultrasound. Circ J. 78:1451–1458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taylor IW, Linding R, Warde-Farley D, Liu
Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q and Wrana JL:
Dynamic modularity in protein interaction networks predicts breast
cancer outcome. Nat Biotechnol. 27:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pujana MA, Han JD, Starita LM, Stevens KN,
Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al:
Network modeling links breast cancer susceptibility and centrosome
dysfunction. Nat Genet. 39:1338–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|