Introduction
Congenital cataracts area common cause of blindness,
with the incidence estimated to be 1–6/10,000 infants in most
populations (1,2). There are 13,000–200,000 patients with
bilateral congenital cataract who go blind each year worldwide,
with an increase of 2,000–40,000 per year (3). The primary clinical manifestation of
the disease is the occurrence of lens opacity in the first year
(4). Although surgical techniques
and visual prognosis have improved, congenital cataracts remain the
leading cause of visual disability in children worldwide (5). Previous studies have revealed that
almost one-third of congenital cataracts are caused by genetic
mutations (6), and 13 genes have
previously been confirmed to be associated with congenital
cataracts (7). These include
crystallin genes [αA-crystallin (CRYAA), αB-crystallin,
βΑ1-crystallin (CRYBA1), βB1-crystallin, βB2-crystallin (CRYBB2),
γC-crystallin and γD-crystallin (CRYGD)], membrane transport
protein genes [major intrinsic protein (MIP), gap junction protein
(GJ)A3 and GJA8], a cytoskeletal protein gene [beaded filament
structural protein 2 (BFSP2)], and transcription factor genes
(paired-like homeodomain 3 and heat shock transcription factor
4).
Evidence indicates that gene expression in the lens
epithelium is significantly altered during cataract formation.
Sheets et al (8) reported the
downregulation of CRYAA and CRYBA1/CRYBA3 and the upregulation of
the receptor tyrosine kinase adhesion-related kinase (ARK) in the
Emory mouse, a well-characterized model of age-dependent cataracts.
Furthermore, metallothionein-IIA, osteonectin and ARK are
upregulated in cataractous lenses relative to transparent lenses
(9–11). Ruotolo et al (12) identified extensive downregulation of
genes, including GCS1, GRB7, FST and POLR2E, in the lens associated
with the development of age-related cataracts in humans. Although
these changes in gene expression are informative, further gene
identifications are required to elucidate the molecular mechanism
of cataract formation. Crystallins, CRYBB2 in particular, are
considered to act primarily as structural proteins of the lens
(13). Previously, it was
demonstrated that the relative amounts of CRYBB2 protein expression
in the lens change markedly, increasing from 12 to 24% (14), suggesting that CRYBB2 serves a
contributive function in lens development. Moreover, targeted
knockout (KO) of CRYBB2 in mice has been demonstrated to induce
age-related (15) and congenital
cataracts (16); however, its
functional significance is not yet known.
Long non-coding RNAs (lncRNAs) are defined as
non-coding RNA molecules >200 nucleotides in length with limited
protein coding potential (17,18).
Previous studies have indicated that lncRNAs are deregulated in
numerous diseases and associated with a wide range of biological
processes, such as proliferation, apoptosis and cell migration
(19,20). Recently, some lncRNAs have been
identified to serve critical functions in eye development and
diseases. Shen et al (21)
reported that 38 lncRNAs were differentially expressed between
transparent and cataractous lenses, among which one of the most
abundant lncRNAs, myocardial infarction associated transcript, was
specifically upregulated in the plasma fraction of whole blood and
the aqueous humor of cataract patients. However, the function of
lncRNAs in human lenses remains unknown.
In the present study, differences in lncRNA and mRNA
expression between the lenses of untreated mice and CRYBB2
KO-induced cataract mice models were evaluated. A total of149
lncRNAs and 803 mRNAs were identified whose expression was
upregulated, while the expression levels of a further 180 lncRNAs
and 732 mRNAs were downregulated in CRYBB2 KO mice lenses. These
findings suggest a potential function for these lncRNAs and mRNAs
in cataract formation.
Materials and methods
Animals
A total of 3 male wild type (WT) and 3 male CRYBB2
KO BALB/c mice (age, 12 weeks old; weight, 25 g) were provided by
in Genious Targeting Laboratory, Inc. (Ronkonkoma, NY, USA)
(22). Mice with targeted disruption
of the CRYBB2 gene were generated at the company by inserting a neo
expression cassette to replace the first and second exons,
preventing the production of a functional transcript from this
locus. Mice were maintained in an animal facility at 25°C, with a
relative humidity of 60–70%, under a 12-h light/dark cycle with
free access to food and water at the Laboratory Animal Center of
the Changhai Hospital, Second Military Medical University
(Shanghai, China). All procedures were carried out in accordance
with the Chinese legislation on the Use and Care of Laboratory
Animals and the ARVO Statement for the Use of Animals in Ophthalmic
and Vision Research (23) and were
approved by the Institutional Animal Care and Use Committee of
Changhai Hospital, Second Military Medical University (Shanghai,
China).
RNA extraction
Following the sacrifice of the mice, the lenses were
collected and RNA was isolated from the lenses of mice using the
Chomczynski method (24) and was
further purified using an RNeasy MinElute Clean-up kit (Qiagen
GmbH, Hilden, Germany). The RNA concentration was measured with a
Nanodrop spectrophotometer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The A260/A280 ratio was 1.8–2.0 and the quality
of the RNA was verified by agarose gel electrophoresis.
Microarray processing
LncRNA and mRNA expression profiling was performed
using the Affymetrix GeneChip Mouse Gene 2.0 ST array (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. Intensities of target hybridization to respective probe
features were detected by laser scanning of the array. First,
quantile normalization of the microarray data of the 3 untreated
and 3 CRYBB2 KO mice was performed. The data was then log2-scale
transformed. Hierarchical clustering of the lncRNA and mRNA
profiles was performed using Cluster 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm)
(25). The normalized expression
values of the lncRNAs and mRNAs were centered on the median before
unsupervised hierarchical clustering was performed. Clustering was
performed with complete linkage and centered Pearson correlation.
To estimate the accuracy of the measurements, the coefficient of
variance for each measured parameter was determined.
Statistical analysis
Statistical analyses were performed with the use of
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). The Significance Analysis of Microarray method was used to
identify significant gene expression changes between CRYBB2 KO mice
and controls (26). P<0.05 was
considered to indicate a statistically significant difference.
Results
Analysis of lncRNA expression patterns
in CRYBB2 KO mice
The lncRNA expression profiles of lens tissues were
compared using unsupervised hierarchical clustering in 3 untreated
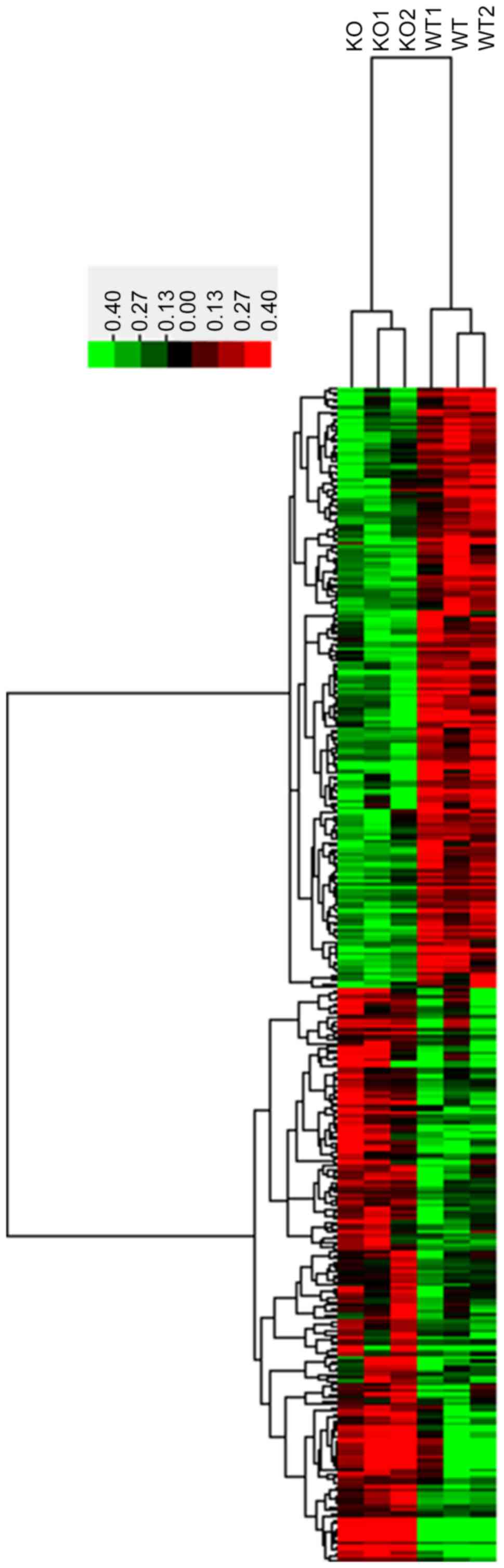
and 3CRYBB2 KO mice. As demonstrated in Fig. 1, in total, 329 lncRNAs with a
coefficient of variance >0.10 were selected for clustering
analysis. Hierarchical clustering of these 329 lncRNAs based on
centered Pearson correlation indicated notable differential lncRNA
expression in CRYBB2 KO and untreated mice (Fig. 1). Among these lncRNAs, 17 exhibited
at least a two-fold change in the CRYBB2 KO mice compared with the
untreated mice (all upregulated in CRYBB2 KO mice). A total of 149
out of 329 lncRNAs were upregulated in CRYBB2 KO mice compared with
the untreated mice (Table I presents
the top 20 most upregulated lncRNAs), whereas 180 out of 329
lncRNAs were downregulated in CRYBB2 KO mice compared with the
untreated mice (Table II presents
the top 20 most downregulated lncRNAs).
 | Table I.Top 20 upregulated long non-coding
RNAs in βB2-crystallin KO mice. |
Table I.
Top 20 upregulated long non-coding
RNAs in βB2-crystallin KO mice.
| Probe set ID | P-value | Fold-change
(KO/WT) | MGI gene symbol | Gene description | GenBank accession
no. |
|---|
| 17296979 |
2.00×10−7 | 5.84 | Gm10409 | Predicted gene
10409 | NR_033121 |
| 17303368 |
1.00×10−6 | 5.21 | Gm3002 | α-takusan
pseudogene | NR_033388 |
| 17296602 |
2.60×10−6 | 3.75 | Gm3020 | Predicted gene
3020 | NR_033117 |
| 17430831 |
3.57×10−5 | 3.55 | Snora73b | Small nucleolar RNA,
H/ACA box 73b | NR_028513 |
| 17303147 |
1.70×10−6 | 3.52 | Gm3591 | Predicted gene
3591 | XR_141206 |
| 17434158 |
6.77×10−3 | 3.51 | – | – |
ENSMUST00000169242 |
| 17337152 |
4.01×10−3 | 3.38 | – | – |
ENSMUST00000174425 |
| 17424407 |
1.21×10−2 | 2.98 | 4933409K07Rik | RIKEN cDNA 4933409K07
gene | NR_033123 |
| 17413061 |
3.08×10−3 | 2.87 | – | – |
ENSMUST00000169242 |
| 17480922 |
3.86×10−4 | 2.68 | Mir139 | MicroRNA139 | NR_029791 |
| 17302054 |
3.04×10−2 | 2.49 | Snora31 | Small nucleolar RNA,
H/ACA box 31 | NR_028481 |
| 17232731 |
3.02×10−2 | 2.4 | Rnu3a | U3A small nuclear
RNA | NR_002842 |
| 17412952 |
9.63×10−3 | 2.32 | Gm3893 | Predicted gene
3893 | NR_033506 |
| 17342996 |
2.00×10−3 | 2.21 | Gm16197 | Predicted gene
16197 | NR_036469 |
| 17421488 |
1.19×10−2 | 2.06 | – | – |
ENSMUST00000172415 |
| 17348121 |
3.41×10−2 | 2.04 | 4833419F23Rik | RIKEN
cDNA4833419F23 gene | NR_040328 |
| 17347279 |
3.47×10−2 | 2.01 | – | – |
ENSMUST00000157334 |
| 17430833 |
1.42×10−3 | 1.98 | Snora73a | Small nucleolar
RNA, H/ACA box 73a | NR_028512 |
| 17221923 |
2.92×10−2 | 1.91 | – | – |
ENSMUST00000083191 |
| 17523680 |
4.81×10−4 | 1.89 | Mir101c | MicroRNA101c | NR_039546 |
 | Table II.Top 20 downregulated long non-coding
RNAs in βB2-crystallin KO mice. |
Table II.
Top 20 downregulated long non-coding
RNAs in βB2-crystallin KO mice.
| Probe set ID | P-value | Fold-change
(KO/WT) | MGI gene
symbol | Gene
description | GenBank accession
no. |
|---|
| 17251898 |
4.62×10−2 | 0.86 | Mir324 | MicroRNA 324 | NR_029758 |
| 17547715 |
4.84×10−2 | 0.86 | – | – |
ENSMUST00000117972 |
| 17468138 |
4.35×10−2 | 0.85 | – | – |
ENSMUST00000145420 |
| 17329209 |
4.58×10−2 | 0.85 | A830060N17 | Uncharacterized
LOC328646 | NR_046162 |
| 17365718 |
3.37×10−2 | 0.84 | – | – |
ENSMUST00000162724 |
| 17403967 |
3.43×10−2 | 0.84 | – | – |
ENSMUST00000158662 |
| 17362668 |
3.62×10−2 | 0.84 | – | – |
ENSMUST00000169060 |
| 17315735 |
3.71×10−2 | 0.84 | – | – |
ENSMUST00000160698 |
| 17527984 |
4.10×10−2 | 0.84 | A730043L09 | Uncharacterized
protein A730043L09 | NR_040769 |
| 17278612 |
4.40×10−2 | 0.84 | Mir342 | MicroRNA342 | NR_029771 |
| 17448958 |
2.17×10−2 | 0.83 | – | – |
ENSMUST00000158856 |
| 17269866 |
3.17×10−2 | 0.83 | – | – |
ENSMUST00000102272 |
| 17225173 |
3.59×10−2 | 0.83 | Snord82 | Small nucleolar
RNA, C/D box 82 | NR_002851 |
| 17230480 |
3.98×10−2 | 0.83 | – | – |
ENSMUST00000128545 |
| 17532275 |
4.53×10−2 | 0.83 | – | – |
ENSMUST00000082463 |
| 17345664 |
4.84×10−2 | 0.83 | – | – |
ENSMUST00000122623 |
| 17280661 |
2.01×10−2 | 0.82 | F730043M19Rik | RIKEN cDNA
F730043M19 gene | NR_015602 |
| 17395003 |
2.17×10−2 | 0.82 | – | – |
ENSMUST00000133525 |
| 17342024 |
2.90×10−2 | 0.82 | Snhg9 | Small nucleolar RNA
host gene (non-protein coding) 9 | NR_027900 |
| 17232800 |
3.09×10−2 | 0.82 | – | – |
ENSMUST00000104610 |
Analysis of mRNA expression patterns
in CRYBB2 KO mice
The mRNA expression profiles of lens tissues were
compared using unsupervised hierarchical clustering in the 3
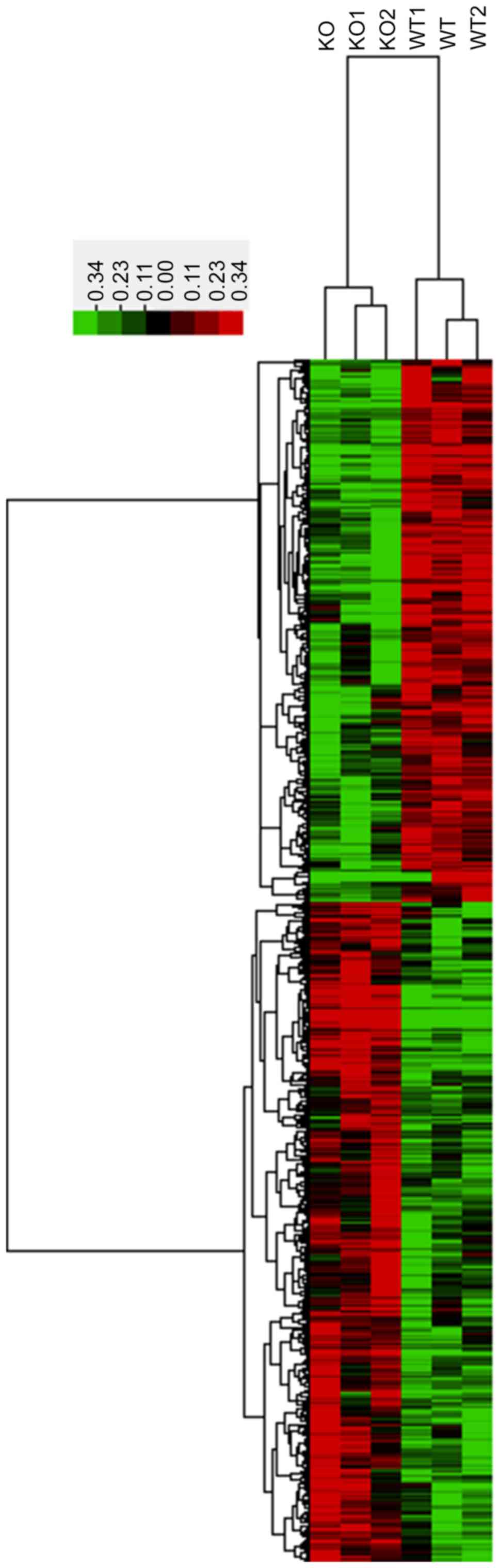
untreated and 3 CRYBB2 KO mice. In total, 1,535 mRNAs with a
coefficient of variance >0.10 were selected for clustering
analysis. Hierarchical clustering of these 1,535 mRNAs based on
centered Pearson correlation indicated notable differential mRNA
expression between CRYBB2 KO and untreated mice (Fig. 2). Among these mRNAs, 52 exhibited at
least a two-fold change in the CRYBB2 KO mice compared with the
untreated mice (all upregulated in CRYBB2 KO mice). A total of 803
out of 1,535 mRNAs were upregulated in CRYBB2 KO mice compared with
the untreated mice (Table III
presents the top 20 most upregulated mRNAs), whereas 732 out of
1,535 mRNAs were downregulated (Table
IV presents the top 20 most downregulated mRNAs).
 | Table III.Top 20 upregulated mRNAs in
βB2-crystallin KO mice. |
Table III.
Top 20 upregulated mRNAs in
βB2-crystallin KO mice.
| Probe set ID | P-value | Fold-change
(KO/WT) | MGI gene
symbol | Gene
description | GenBank accession
no. |
|---|
| 17477347 |
3.60×10−2 | 7.22 | Klk1b22 | Kallikrein
1-related peptidase b22 | NM_010114 |
| 17303018 |
1.00×10−7 | 7.14 | Gm3500 | Predicted gene
3500 | NM_001256886 |
| 17296943 |
2.52×10−4 | 5.78 | – | – |
ENSMUST00000163719 |
| 17303117 |
1.00×10−7 | 5.58 | Gm3696 | Predicted gene
3696 |
ENSMUST00000167923 |
| 17296896 |
4.00×10−7 | 5.14 | Gm5796 | Predicted gene
5796 | NM_001029930 |
| 17548311 |
1.22×10−5 | 4.56 | Gm3579 | Predicted gene
3579 | AY140896 |
| 17373996 |
1.35×10−4 | 4.53 | BC048594 | cDNA sequence
BC048594 | BC048594 |
| 17331088 |
4.22×10−5 | 4.48 | Gm19797 | Predicted gene
19797 | XM_003085996 |
| 17303024 |
3.66×10−4 | 4.13 | Gm10021 | Predicted gene
10021 | AK084071 |
| 17413352 |
5.14×10−3 | 3.46 | Car9 | Carbonic anhydrase
9 |
ENSMUST00000030183 |
| 17296849 |
9.00×10−6 | 3.4 | Gm2897 | Predicted gene
2897 | NM_001177715 |
| 17335467 |
1.21×10−2 | 3.22 | Cdkn1a | Cyclin-dependent
kinase inhibitor 1A (P21) | NM_007669 |
| 17412962 |
7.94×10−3 | 3.2 | Gm3893 | Predicted gene
3893 | BC059060 |
| 17496857 |
2.59×10−2 | 3.16 | Cox6a2 | Cytochrome c
oxidase, subunit VI a, polypeptide 2 | NM_009943 |
| 17466743 |
9.47×10−4 | 3.13 | Npvf | Neuropeptide VF
precursor |
ENSMUST00000031853 |
| 17331078 |
3.77×10−4 | 3.06 | Tmem45a | Transmembrane
protein 45a | NM_019631 |
| 17296836 |
2.00×10−6 | 3.01 | Gm5458 | Predicted gene
5458 | NM_001024706 |
| 17280292 |
5.05×10−4 | 2.93 | – | – |
ENSMUST00000169148 |
| 17296595 |
2.52×10−5 | 2.9 | D830030K20Rik | RIKEN cDNA
D830030K20 gene |
ENSMUST00000169218 |
| 17303315 |
5.57×10−5 | 2.78 | Gm5797 | Predicted gene
5797 |
ENSMUST00000100886 |
 | Table IV.Top 20 downregulated mRNAs in
βB2-crystallin KO mice. |
Table IV.
Top 20 downregulated mRNAs in
βB2-crystallin KO mice.
| Probe set ID | P-value | Fold-change
(KO/WT) | MGI gene
symbol | Gene
description | GenBank accession
no. |
|---|
| 17256388 |
3.86×10−2 | 0.86 | Ttc25 | Tetratricopeptide
repeat domain 25 | NM_028918 |
| 17283203 |
4.91×10−2 | 0.86 | Foxn3 | Forkhead box
N3 |
ENSMUST00000046859 |
| 17357502 |
4.94×10−2 | 0.86 | Cpsf7 | Cleavage and
polyadenylation specific factor 7 | NM_172302 |
| 17440775 |
4.31×10−2 | 0.86 | Dao | D-amino acid
oxidase |
ENSMUST00000112292 |
| 17473796 |
4.34×10−2 | 0.86 | Rps5 | Ribosomal protein
S5 | NM_009095 |
| 17528663 |
4.79×10−2 | 0.86 | Polr2m | Polymerase (RNA) II
(DNA directed) polypeptide M | NM_178602 |
| 17231003 |
4.52×10−2 | 0.85 | Mfsd7b | Major facilitator
superfamily domain containing 7B | NM_001081259 |
| 17219005 |
4.68×10−2 | 0.85 | Creg1 | Cellular repressor
of E1A-stimulated genes 1 | NM_011804 |
| 17225580 |
4.98×10−2 | 0.85 | Olfr1415 | Olfactory receptor
1415 | NM_001011525 |
| 17256716 |
4.83×10−2 | 0.85 | Rundc1 | RUN domain
containing 1 | NM_172566 |
| 17291143 |
3.31×10−2 | 0.85 | – | – | AK029074 |
| 17304186 |
4.98×10−2 | 0.85 | Plac9 | Placenta specific
9 | NM_207229 |
| 17358640 |
4.57×10−2 | 0.85 | Mbl2 | Mannose-binding
lectin (protein C) 2 |
ENSMUST00000025797 |
| 17368499 |
4.89×10−2 | 0.85 | Dbh | Dopamine beta
hydroxylase |
ENSMUST00000000910 |
| 17425160 |
3.58×10−2 | 0.85 | Erp44 | Endoplasmic
reticulum protein 44 | NM_029572 |
| 17451345 |
4.98×10−2 | 0.85 | 2900026A02Rik | RIKEN cDNA
2900026A02 gene | NM_172884 |
| 17501800 |
3.53×10−2 | 0.85 | Hapln4 | Hyaluronan and
proteoglycan link protein 4 | NM_177900 |
| 17499224 |
3.82×10−2 | 0.85 | F10 | Coagulation factor
X | NM_001242368 |
| 17504309 |
3.96×10−2 | 0.85 | Ccdc113 | Coiled-coil domain
containing 113 | NM_172914 |
| 17502071 |
4.01×10−2 | 0.85 | – | – |
ENSMUST00000050921 |
Discussion
In the present study, the lncRNA and mRNA profiles
of untreated and CRYBB2 KO cataractous lenses were evaluated. A
total of 149 lncRNAs and 803 mRNAs were identified to be
upregulated, while 180 lncRNAs and 732 mRNAs were identified to be
downregulated in CRYBB2 KO mice lenses, implying a potential role
of these lncRNAs and mRNAs in cataract formation.
In previous research, an increasing number of
lncRNAs have been identified and associations between lncRNAs and
numerous diseases, including cardiovascular and neurodegeneration
diseases, have been reported (27).
The roles of lncRNAs in cancer development are being studied
(28,29). However, the function of lncRNAs in
disease, particularly in cataracts, has not yet been reported. To
the best of our knowledge, the current study presents the first
report on differential lncRNA expression in a cohort of mice with
or without CRYBB2 KO. Through an analysis of lenses, it was
identified that 329 lncRNAs were differentially expressed in CRYBB2
KO and untreated mice, suggesting that lncRNAs may serve critical
functions in cataract formation. Among the top 20 most upregulated
lncRNAs, five were predicted genes and a further six were unnamed.
Among the top 20 most downregulated lncRNAs, 13 lncRNAs were
unnamed and the others were known (identified with Mouse Genome
Informatics gene symbols). These results indicated that these
lncRNAs were linked with CRYBB2-associated cataract formation.
Notably, the expression changes of lncRNAs in the upregulated group
(maximum change, 5.84-fold) were higher compared with those in the
downregulated group (maximum change, 0.86-fold), suggesting a
higher susceptibility of lncRNAs to be upregulated rather than
downregulated in cataracts.
Differential mRNA expression was also examined in
the cohort of mice with or without CRYBB2 KO. Through an analysis
of lenses, it was identified that 1,535 mRNAs were differentially
expressed between CRYBB2 KO and untreated mice. Among the top 20
most upregulated mRNAs, 10 mRNAs were predicted genes and two mRNAs
were unnamed. These results indicated that these mRNAs may serve
critical functions in cataract formation. Notably, the expression
changes of mRNAs in the upregulated group (maximum change,
7.22-fold) were higher compared with those in the downregulated
group (maximum change, 0.86-fold), suggesting a higher
susceptibility of mRNAs to be upregulated rather than downregulated
in cataracts.
A previous limited microarray survey with a panel of
cell cycle-regulated genes illustrated that irradiation with
protons altered the gene expression pattern of human lens
epithelial cells (30), such as
cyclin-dependent kinase inhibitor 1 (CDKN1A), which codes for a
protein that is involved in several pathways functionally
associated with linear energy transfer-responsive radiation damage.
Cytochrome C oxidase 6A2 (COX6A2) was identified to be upregulated
during cataract development in mice with a mutation in MIP, a
functional water channel that serves a key role in establishing
lens fiber cell architecture and is associated with inherited and
age-related forms of cataracts (31). Consistent with these results, the
present study also identified upregulated expression of CDKN1A and
COX6A2 in a CRYBB2 KO-induced cataract mice model. Furthermore,
Fas-mediated apoptosis in human lens epithelial cells of cataracts
is associated with diabetic retinopathy (32), suggesting a role for Fas in cataract
formation, which is contrary to the finding of the present study
that Fas was downregulated in a CRYBB2 KO-induced cataracts mouse
model (data not shown). BFSP2, a gene for a lens-specific beaded
filament structural protein, was down-regulated in CRYBB2
KO-induced cataract mice (33,34),
which is in agreement with the findings of the present study: BFSP2
expression is restricted to the lens fiber cells, and a deletion
mutation of BFSP2 is associated with cataracts. CRYGD mutation has
previously been observed to cause autosomal dominant congenital
cerulean cataracts, suggesting an inhibitory role of CRYGD in
cataract formation (35). This is
consistent with the current findings that CRYGD is downregulated in
a CRYBB2 KO-induced cataracts mouse model (data not shown).
The present study has some limitations, including
the relatively small number of mice in each cohort, and the fact
that only RNA samples from the lens were utilized for
hybridizations. Furthermore, the differentially expressed lncRNAs
and mRNAs require further clarification in future
investigations.
In conclusion, knowledge of the changes in lncRNA
and mRNA expression associated with cataracts may contribute to a
better understanding of the opacification process. The findings of
the present study demonstrate that there are notable lncRNA and
mRNA differences between mice with or without CRYBB2 KO induction.
The data indicate that the response of the lens to the development
of CRYBB2 KO-related cataract is characterized by an extensive
upregulation of numerous mRNAs and lncRNAs.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Science Foundation of China (grant nos. 81300748 and 81170834).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YJ conceived of and designed the experiments. KX,
H-XR and W-JL performed the experiments and analyzed the data. W-JL
and KX obtained the reagents, materials and analysis tools. YJ and
W-JL wrote the study. All authors read and approved the final
study.
Ethics approval and consent to
participate
All procedures were carried out in accordance with
the Chinese legislation on the Use and Care of Laboratory Animals
and the ARVO Statement for the Use of Animals in Ophthalmic and
Vision Research (23) and were
approved by the Institutional Animal Care and Use Committee of
Changhai Hospital, Second Military Medical University (Shanghai,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reddy MA, Francis PJ, Berry V,
Bhattacharya SS and Moore AT: Molecular genetic basis of inherited
cataract and associated phenotypes. Surv Ophthalmol. 49:300–315.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holmes JM, Leske DA, Burke JP and Hodge
DO: Birth prevalence of visually significant infantile cataract in
a defined US population. Ophthalmic Epidemiol. 10:67–74. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foster A, Gilbert C and Rahi J:
Epidemiology of cataract in childhood: A global perspective. J
Cataract Refract Surg. 23 Suppl 1:S601–S604. 1997. View Article : Google Scholar
|
|
4
|
Peng CH, Liu JH, Woung LC, Lin TJ, Chiou
SH, Tseng PC, Du WY, Cheng CK, Hu CC, Chien KH and Chen SJ:
MicroRNAs and cataracts: Correlation among let-7 expression, age
and the severity of lens opacity. Br J Ophthalmol. 96:747–751.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
You C, Wu X, Zhang Y, Dai Y, Huang Y and
Xie L: Visual impairment and delay in presentation for surgery in
chinese pediatric patients with cataract. Ophthalmology. 118:17–23.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao X, Cheng J, Lu C, Li X, Li F, Liu C,
Zhang M, Zhu S and Ma X: A novel mutation in the connexin 50 gene
(GJA8) associated with autosomal dominant congenital nuclear
cataract in a Chinese family. Curr Eye Res. 35:597–604. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen C, Sun Q, Gu M, Liu K, Sun Y and Xu
X: A novel Cx50 (GJA8) p. H277Y mutation associated with autosomal
dominant congenital cataract identified with targeted
next-generation sequencing. Graefes Arch Clin Exp Ophthalmol.
253:915–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheets NL, Chauhan BK, Wawrousek E,
Hejtmancik JF, Cvekl A and Kantorow M: Cataract-and lens-specific
upregulation of ARK receptor tyrosine kinase in Emory mouse
cataract. Invest Ophthalmol Vis Sci. 43:1870–1875. 2002.PubMed/NCBI
|
|
9
|
Hawse JR, Padgaonkar VA, Leverenz VR,
Pelliccia SE, Kantorow M and Giblin FJ: The role of metallothionein
IIa in defending lens epithelial cells against cadmium and TBHP
induced oxidative stress. Mol Vis. 12:342–349. 2006.PubMed/NCBI
|
|
10
|
Gilmour DT, Lyon GJ, Carlton MB, Sanes JR,
Cunningham JM, Anderson JR, Hogan BL, Evans MJ and Colledge WH:
Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40
develop normally but show severe age-onset cataract formation and
disruption of the lens. EMBO J. 17:1860–1870. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hawse JR, Hejtmancik JF, Horwitz J and
Kantorow M: Identification and functional clustering of global gene
expression differences between age-related cataract and clear human
lenses and aged human lenses. Exp Eye Res. 79:935–940. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruotolo R, Grassi F, Percudani R, Rivetti
C, Martorana D, Maraini G and Ottonello S: Gene expression
profiling in human age-related nuclear cataract. Mol Vis.
9:538–548. 2003.PubMed/NCBI
|
|
13
|
Pauli S, Söker T, Klopp N, Illig T, Engel
W and Graw J: Mutation analysis in a German family identified a new
cataract-causing allele in the CRYBB2 gene. Mol Vis. 13:962–967.
2007.PubMed/NCBI
|
|
14
|
Lou D, Tong J, Zhang L, Chiang SW, Lam DS
and Pang C: A novel mutation in CRYBB2 responsible for inherited
coronary cataract. Eye (Lond). 23:1213–1220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Li J, Huang C, Xue L, Peng Y, Fu
Q, Gao L, Zhang J and Li W: Targeted knockout of the mouse
betaB2-crystallin gene (Crybb2) induces age-related cataract.
Invest Ophthalmol Vis Sci. 49:5476–5483. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mothobi ME, Guo S, Liu Y, Chen Q, Yussuf
AS, Zhu X and Fang Z: Mutation analysis of congenital cataract in a
Basotho family identified a new missense allele in CRYBB2. Mol Vis.
15:1470–1475. 2009.PubMed/NCBI
|
|
17
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Ann Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen Y, Dong LF, Zhou RM, Yao J, Song YC,
Yang H, Jiang Q and Yan B: Role of long non-coding RNA MIAT in
proliferation, apoptosis and migration of lens epithelial cells: A
clinical and in vitro study. J Cell Mol Med. 20:537–548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Huang C, Li W, Wang J and Weng W:
Establishment of a βB2 crystallin gene knockout mice model. Acad J
Second Mil Med Univ. 27:1246–1249. 2006.
|
|
23
|
Delgado D, del Pozo-Rodríguez A, Solinís
MÁ, Avilés-Triqueros M, Weber BH, Fernández E and Gascón AR:
Dextran and protamine-based solid lipid nanoparticles as potential
vectors for the treatment of X-linked juvenile retinoschisis. Hum
Gene Ther. 23:345–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chomczynski P and Sacchi N: The
single-step method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction: Twenty-something years
on. Nat Protoc. 1:581–585. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41:(Database issue). D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang PY, Bjornstad KA, Rosen CJ, McNamara
MP, Mancini R, Goldstein LE, Chylack LT and Blakely EA: Effects of
iron ions, protons and X rays on human lens cell differentiation.
Radiat Res. 164:531–539. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Bennett TM and Shiels A: Lens
ER-stress response during cataract development in Mip-mutant mice.
Biochim Biophys Acta. 1862:1433–1442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Okamura N, Ito Y, Shibata MA, Ikeda T and
Otsuki Y: Fas-mediated apoptosis in human lens epithelial cells of
cataracts associated with diabetic retinopathy. Med Electron
Microsc. 35:234–241. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jakobs PM, Hess JF, FitzGerald PG, Kramer
P, Weleber RG and Litt M: Autosomal-dominant congenital cataract
associated with a deletion mutation in the human beaded filament
protein gene BFSP2. Am J Hum Genet. 66:1432–1436. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang L, Gao L, Li Z, Qin W, Gao W, Cui X,
Feng G, Fu S, He L and Liu P: Progressive sutural cataract
associated with a BFSP2 mutation in a Chinese family. Mol Vis.
12:1626–1631. 2006.PubMed/NCBI
|
|
35
|
Santana A, Waiswol M, Arcieri ES, de
Vasconcellos Cabral JP and de Melo Barbosa M: Mutation analysis of
CRYAA, CRYGC, and CRYGD associated with autosomal dominant
congenital cataract in Brazilian families. Mol Vis. 15:793–800.
2009.PubMed/NCBI
|