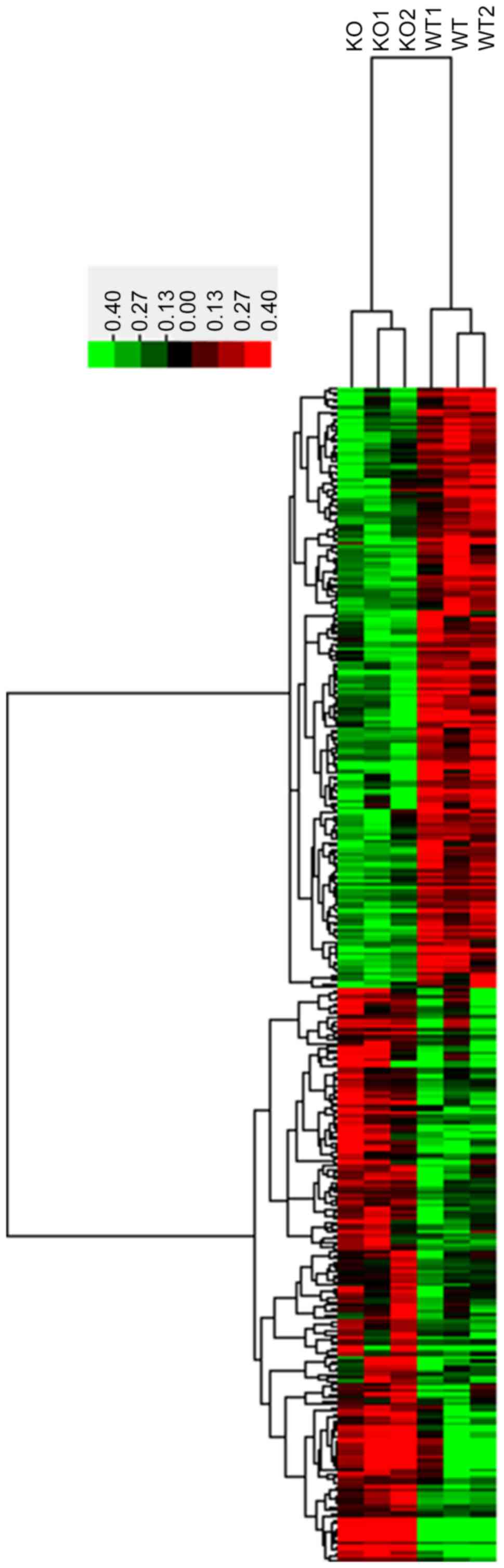
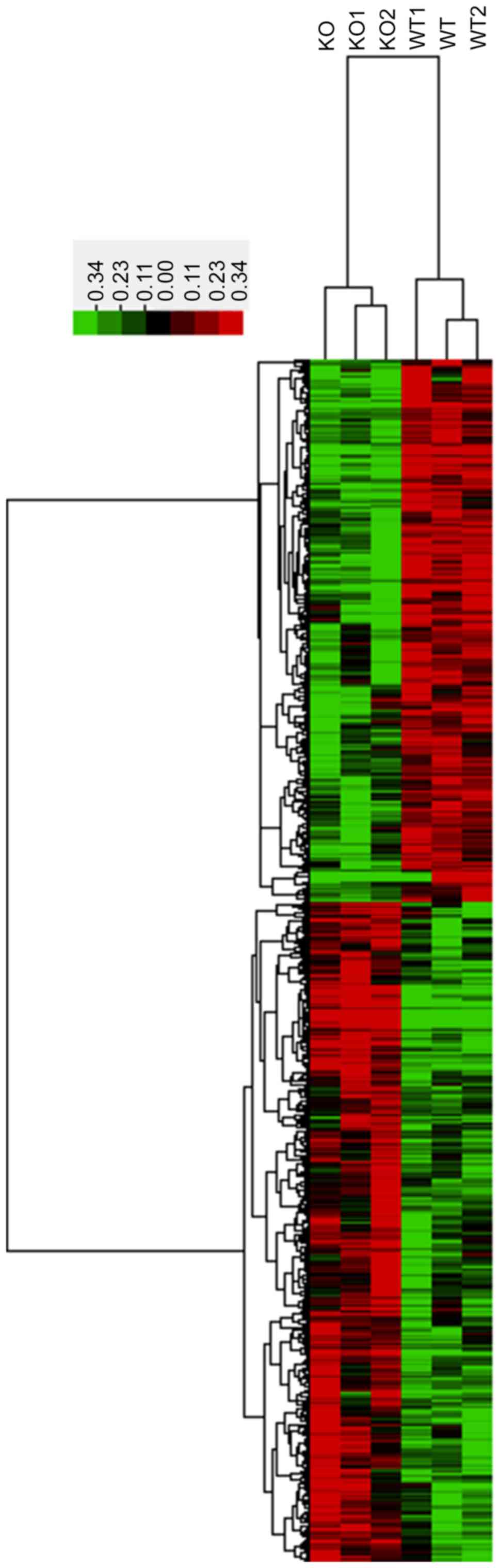
|
1
|
Reddy MA, Francis PJ, Berry V,
Bhattacharya SS and Moore AT: Molecular genetic basis of inherited
cataract and associated phenotypes. Surv Ophthalmol. 49:300–315.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holmes JM, Leske DA, Burke JP and Hodge
DO: Birth prevalence of visually significant infantile cataract in
a defined US population. Ophthalmic Epidemiol. 10:67–74. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foster A, Gilbert C and Rahi J:
Epidemiology of cataract in childhood: A global perspective. J
Cataract Refract Surg. 23 Suppl 1:S601–S604. 1997. View Article : Google Scholar
|
|
4
|
Peng CH, Liu JH, Woung LC, Lin TJ, Chiou
SH, Tseng PC, Du WY, Cheng CK, Hu CC, Chien KH and Chen SJ:
MicroRNAs and cataracts: Correlation among let-7 expression, age
and the severity of lens opacity. Br J Ophthalmol. 96:747–751.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
You C, Wu X, Zhang Y, Dai Y, Huang Y and
Xie L: Visual impairment and delay in presentation for surgery in
chinese pediatric patients with cataract. Ophthalmology. 118:17–23.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao X, Cheng J, Lu C, Li X, Li F, Liu C,
Zhang M, Zhu S and Ma X: A novel mutation in the connexin 50 gene
(GJA8) associated with autosomal dominant congenital nuclear
cataract in a Chinese family. Curr Eye Res. 35:597–604. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen C, Sun Q, Gu M, Liu K, Sun Y and Xu
X: A novel Cx50 (GJA8) p. H277Y mutation associated with autosomal
dominant congenital cataract identified with targeted
next-generation sequencing. Graefes Arch Clin Exp Ophthalmol.
253:915–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheets NL, Chauhan BK, Wawrousek E,
Hejtmancik JF, Cvekl A and Kantorow M: Cataract-and lens-specific
upregulation of ARK receptor tyrosine kinase in Emory mouse
cataract. Invest Ophthalmol Vis Sci. 43:1870–1875. 2002.PubMed/NCBI
|
|
9
|
Hawse JR, Padgaonkar VA, Leverenz VR,
Pelliccia SE, Kantorow M and Giblin FJ: The role of metallothionein
IIa in defending lens epithelial cells against cadmium and TBHP
induced oxidative stress. Mol Vis. 12:342–349. 2006.PubMed/NCBI
|
|
10
|
Gilmour DT, Lyon GJ, Carlton MB, Sanes JR,
Cunningham JM, Anderson JR, Hogan BL, Evans MJ and Colledge WH:
Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40
develop normally but show severe age-onset cataract formation and
disruption of the lens. EMBO J. 17:1860–1870. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hawse JR, Hejtmancik JF, Horwitz J and
Kantorow M: Identification and functional clustering of global gene
expression differences between age-related cataract and clear human
lenses and aged human lenses. Exp Eye Res. 79:935–940. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruotolo R, Grassi F, Percudani R, Rivetti
C, Martorana D, Maraini G and Ottonello S: Gene expression
profiling in human age-related nuclear cataract. Mol Vis.
9:538–548. 2003.PubMed/NCBI
|
|
13
|
Pauli S, Söker T, Klopp N, Illig T, Engel
W and Graw J: Mutation analysis in a German family identified a new
cataract-causing allele in the CRYBB2 gene. Mol Vis. 13:962–967.
2007.PubMed/NCBI
|
|
14
|
Lou D, Tong J, Zhang L, Chiang SW, Lam DS
and Pang C: A novel mutation in CRYBB2 responsible for inherited
coronary cataract. Eye (Lond). 23:1213–1220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Li J, Huang C, Xue L, Peng Y, Fu
Q, Gao L, Zhang J and Li W: Targeted knockout of the mouse
betaB2-crystallin gene (Crybb2) induces age-related cataract.
Invest Ophthalmol Vis Sci. 49:5476–5483. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mothobi ME, Guo S, Liu Y, Chen Q, Yussuf
AS, Zhu X and Fang Z: Mutation analysis of congenital cataract in a
Basotho family identified a new missense allele in CRYBB2. Mol Vis.
15:1470–1475. 2009.PubMed/NCBI
|
|
17
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Ann Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen Y, Dong LF, Zhou RM, Yao J, Song YC,
Yang H, Jiang Q and Yan B: Role of long non-coding RNA MIAT in
proliferation, apoptosis and migration of lens epithelial cells: A
clinical and in vitro study. J Cell Mol Med. 20:537–548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Huang C, Li W, Wang J and Weng W:
Establishment of a βB2 crystallin gene knockout mice model. Acad J
Second Mil Med Univ. 27:1246–1249. 2006.
|
|
23
|
Delgado D, del Pozo-Rodríguez A, Solinís
MÁ, Avilés-Triqueros M, Weber BH, Fernández E and Gascón AR:
Dextran and protamine-based solid lipid nanoparticles as potential
vectors for the treatment of X-linked juvenile retinoschisis. Hum
Gene Ther. 23:345–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chomczynski P and Sacchi N: The
single-step method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction: Twenty-something years
on. Nat Protoc. 1:581–585. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41:(Database issue). D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang PY, Bjornstad KA, Rosen CJ, McNamara
MP, Mancini R, Goldstein LE, Chylack LT and Blakely EA: Effects of
iron ions, protons and X rays on human lens cell differentiation.
Radiat Res. 164:531–539. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Bennett TM and Shiels A: Lens
ER-stress response during cataract development in Mip-mutant mice.
Biochim Biophys Acta. 1862:1433–1442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Okamura N, Ito Y, Shibata MA, Ikeda T and
Otsuki Y: Fas-mediated apoptosis in human lens epithelial cells of
cataracts associated with diabetic retinopathy. Med Electron
Microsc. 35:234–241. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jakobs PM, Hess JF, FitzGerald PG, Kramer
P, Weleber RG and Litt M: Autosomal-dominant congenital cataract
associated with a deletion mutation in the human beaded filament
protein gene BFSP2. Am J Hum Genet. 66:1432–1436. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang L, Gao L, Li Z, Qin W, Gao W, Cui X,
Feng G, Fu S, He L and Liu P: Progressive sutural cataract
associated with a BFSP2 mutation in a Chinese family. Mol Vis.
12:1626–1631. 2006.PubMed/NCBI
|
|
35
|
Santana A, Waiswol M, Arcieri ES, de
Vasconcellos Cabral JP and de Melo Barbosa M: Mutation analysis of
CRYAA, CRYGC, and CRYGD associated with autosomal dominant
congenital cataract in Brazilian families. Mol Vis. 15:793–800.
2009.PubMed/NCBI
|