Introduction
Donor organ rejection remains a significant problem
for patients receiving transplants (1,2).
Pharmacological suppression of immune responses can suppress the
rejection process, but the resultant nonspecific suppression of
immune responses can lead to a variety of problems, including tumor
and infection (3,4). To improve graft survival rates and
eliminate the requirement for immunosuppression, donor-specific
transplant tolerance must be achieved (5,6).
Therefore, a treatment protocol in which the organ recipient is
induced to recognize the donor tissue as ‘self’ would be highly
desirable.
The major histocompatibility complex (MHC) is the
principal system responsible for recognizing transplanted
allogeneic or heterogeneous tissues by the immune system (7,8). The MHC
is composed of a series of genes and produces two classes of cell
surface glycoproteins and specific serum proteins (MHC class I, II
and III molecules) (9,10). Since a discrepancy between the
donor's MHC antigen and that of the recipient leads to rejection
(10), improving MHC compatibility
is desirable (11). It was
hypothesized that if the donor's MHC genes could be introduced into
and expressed by the recipient, tolerance may be induced.
T cells serve a key role in the immune system and in
transplantation rejection. Following the development of the cells
in the bone marrow, T cells undergo positive and negative
selection, interact with MHC antigen in the thymus, then
differentiate and mature (12–14).
Through the process of negative selection, T cells that react with
self-MHC antigen are removed by apoptosis (15). It was therefore surmised that if the
donor's MHC were transferred into a recipient's thymus, apoptosis
would also be induced in T cells that react to the expressed donor
MHC antigen.
In mice, MHC is also termed mouse
histocompatibility-2 complex(H-2). This gene complex includes K, D,
L, I and S loci. The K, D and L loci code for the MHC class I
antigen, which is expressed on the surface of various types of
cells and causes strong rejection following transplantation. The I
region, which includes Aα, Aβ, Eα and Eβ loci, codes for the MHC
class II antigen, which is expressed on the surface of immune cells
(12). The S locus is MHC class III
gene that codes for specific serum proteins (12). The K locus is the most important of
the three MHC class I loci and antigens of this class are present
on the surface of nearly all cells in the mouse (16). For these reasons, a previous study by
our group demonstrated that the rejection of transplanted hearts
was mitigated and the survival time of transplanted hearts were
prolonged following the transfer of a mouse donor's K locus into a
mouse recipient's thymus (17).
However, since the K locus is only a part of the MHC gene complex,
the present study aimed to investigate whether transferring all of
a donor's loci of the MHC I and II genes into a recipient would be
more effective in mitigating rejection and improving graft survival
time. In addition, to examine heterograft effects, mouse donor K
loci were also transferred into rats.
Materials and methods
Animals
Experiments were performed using a total of 45 6- to
8-week-old male and female mice (weight 20–25 g) and a total of 20
8- to 10-week-old male and female rats (weight, 200–300 g). The
animals (Charles River Laboratories, Beijing, China) were fed in
the animal lab of Capital Medical University and maintained at
20–26°C with a level 7 air cleanliness, a 12 h light/dark cycle and
daily access to food and water. Balb/c mice (the haploid was
H-2d) were used as donors, and C57BL/6 mice
(H-2b) and Sprague Dawley rats were used as recipients.
All experiments were approved by the Capital Medical University
Institutional Animal Care and Use Committee (Beijing, China).
Materials
AMV reverse transcriptase and pCI-neo vector were
purchased from Promega Corporation (Madison, WI, USA). Mouse
anti-H-2d-K (cat. no. BE0104; clone HB159) and
anti-H-2d-D (cat. no. BE0180; clone 34-1-2S) antibodies
were purchased from Bio X Cell (West Lebanon, NH, USA). The MTT kit
was purchased from Fitzgerald Industries International (North
Acton, MA, USA). The primers were obtained from Invitrogen (Thermo
Fisher Scientific, Inc., Waltham, USA).
Preparation of donor H-2d
gene
Total RNA was extracted from the donors' livers
using an RNAprep pure kit (for Tissue) (DP431) (Tiangen Biotech
Co., Ltd., Beijing, China) and then reverse transcribed to obtain
cDNA using AMV reverse transcriptase (RT) and an AMV reverse
transcriptase buffer (10 mM dNTP mixture); the two experiments were
performed according to the manufacturer's protocol at 42°C for 60
min. The donor H-2d gene was amplified by nested
polymerase chain reaction (PCR). For the first PCR step, the
primers were designed according to the 5′ and 3′ untranslated
region in mRNA of every locus. cDNA served as the templates. The
PCR protocol included pre-degeneration for 3 min at 94°C, followed
by 30 cycles of degeneration at 94°C for 30 sec, annealing at 55°C
for 30 sec, and elongation at 72°C for 2 min. As an internal
reference, mouse β-actin mRNA was amplified using primers
5′-CCCCATTGAACATGGCATTG-3′ and 5′-ACGACCAGAGGCATACAGG-3′. The
reaction was run using Taq Platinum PCR MasterMix or pfu MasterMix
(Tiangen Biotech Co., Ltd.).
The second PCR analysis was performed using the
product of the first PCR as the template. The primers were designed
according to the 5′ and 3′ end of the coding sequence in mRNA of
every locus. The primers are indicated in Table I. The PCR protocol was identical to
that used in the first step. The product of the second PCR step was
retrieved by electrophoresis on a 1% agarose gel (visualized by 0.5
µg/ml ethidium bromide) and verified by sequence analysis using
NextGENe 2.3 (SoftGenetics, LLC, State College, PA, USA).
 | Table I.Primers sequences used in polymerase
chain reaction analysis. |
Table I.
Primers sequences used in polymerase
chain reaction analysis.
|
| Primers of UTR
(5′-3′) | Primers of CDS
(5′-3′) |
|---|
|
|
|
|
|---|
| Target gene | Forward | Reverse | Forward | Reverse |
|---|
|
H-2Kd |
CGCTAATCGCCGACCAGT |
TGAGGGCTCTGGATGTCACA |
ATGGCACCCTGCACGCT |
TCACGCTAGAGAATGAGGGTCA |
|
H-2Dd |
GCGGACGCTGGTTATAAAGTC |
GCAGACTCACAGGGAACATCAG |
ATGGGGGCGATGGCTCCG |
TCACACTTTACAATCTGGGAGAGACAT |
|
H-2Ld |
ATCCCAGATGGGGGCGAT |
GGCTCTGGATGTCACAGGAGA |
ATGGGGGCGATGGCTCC |
TCACGCTTTACAATCTCGGAGA |
|
H-2Aαd |
GCAGAGACCTCCCAGAGACC |
ACCTTCCTTTCCAGGGTGTG |
ATGCCGTGCAGCAGAGCT |
TCATAAAGGCCCTGGGTGTC |
|
H-2Aβd |
TCCTGGTGACTGGCATTACCT |
CATGCAGGCCTTACAGTCTGA |
ATGGCTCTGCAGATCCCC |
TCACTGCAGGAGCCCTGC |
|
H-2Eαd |
GCTTCTGAACCCACCAAACA |
GAAGGCATTGCCTCCAGGTA |
ATGGCCACAATTGGAGCC |
TCACAGGGCTCCTTGTCGG |
|
H-2Eβd |
TTCCCCTCTGACTCCTGTGTC |
ACTCCTTCCTTCAGCCTTGTTAC |
ATGGTGTGGCTCCCCAGAG |
TCAGCTCAGGAGTCCTGTTGG |
Construction of the expression
vector
The retrieved H-2 DNA fragments of each locus, K, D,
L, Aα, Aβ, Eα and Eβ were cloned into the pBS-T vector (50 ng/µl;
Tiangen Biotech Co., Ltd.). Escherichia coli (E.
coli) TOP-10 cells (Tiangen Biotech Co., Ltd.) were transformed
using recombinant DNA and vector. The competent TOP-10 cells were
combined with recombinant DNA and the vector on ice for 20 min,
heated to 42°C for 90 sec, put on ice again for 3 min, added to the
Luria-Bertani (LB) medium (Tiangen Biotech Co., Ltd.) and agitated
for 1 h at 37°C. Blue-white screening (with Ampicillin, 100 mg/l in
LB medium) and PCR (by the same protocol as the second PCR above)
were performed to select clones. Selected colonies were then
cultivated until the cells were harvested and the plasmid was
extracted. Following this, recombinant DNA digested with
endonucleases Xho1 and EcoR1, and ligated into
pCI-neo (all Promega Corporation), yielding
pCI-neo-H-2Kd, pCI-neo-H-2Dd,
pCI-neo-H-2Ld, pCI-neo-H-2Aαd,
pCI-neo-H-2Aβd, pCI-neo-H-2Eαd and
pCI-neo-H-2Eβd. All the aforementioned recombinant DNA
were cloned into E. coli TOP-10 cells as above.
Thymus injection and heart
transplantation
A total of 45 recipient mice were randomly divided
into three groups (n=15/group) and received plasmid injections with
pCI-neo-H-2Kd, seven vectors with all seven loci of the
H-2d gene or an empty vector, which was used as a
control. A total of 20 recipient rats were randomly divided
(n=10/group) to receive either pCI-neo-H-2Kd or empty
vector, which was used as a control. The mouse and rat recipients
were first anesthetized using 5% chloral hydrate (Tiangen Biotech
Co., Ltd.) at 350 mg/kg. The mice and rats stopped moving following
the administration of anesthesia, but their eyes were usually
opened. Palpebral reflex, toe pinch reflex and corneal reflex tests
were performed to monitor the depth of anesthesia. Using an
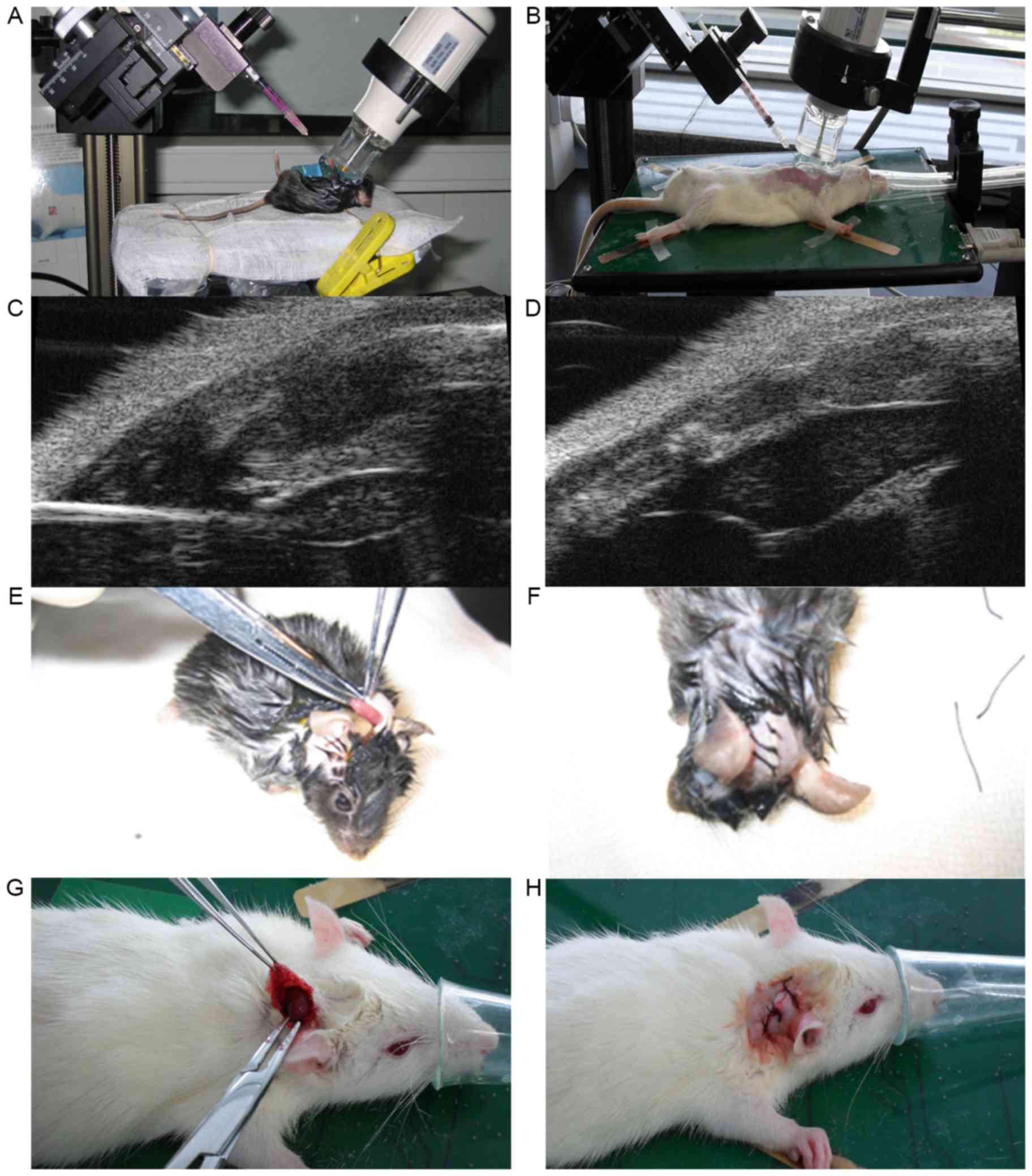
ultrasound for guidance, the thymi of the animals were punctured
(Fig. 1A-D). All recipients in the
mouse and rat control groups were injected with 0.7 µg empty
pCI-neo vectors + 1.4 µl lipofectin (Invitrogen, Thermo Fisher
Scientific, Inc.) [diluted with Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.) up to 70 µl].
The1-vector recipient mice were injected with 0.1 µg
pCI-neo-H-2Kd + 0.6 µg empty pCI-neo vectors + 1.4 µl
lipofectin (diluted with DMEM up to 70 µl). The 7-vector recipient
mice were injected with 0.1 µg pCI-neo-H-2Kd + 0.1 µg
pCI-neo-H-2Dd + 0.1 µg pCI-neo-H-2Ld + 0.1 µg
pCI-neo-H-2Aαd + 0.1 µg pCI-neo-H-2Aβd + 0.1
µg pCI-neo-H-2Eαd + 0.1 µg pCI-neo-H-2Eβd +
1.4 µl lipofectin (diluted with DMEM up to 70 µl). Recipient rats
of the 1-vector experimental group were injected with 0.7 µg
pCI-neo-H-2Kd + 1.4 µl lipofectin (diluted by DMEM up to
70 µl).
Heart transplantations were performed immediately
following the injections using the ear-back heart transplantation
model (18,19). The donor hearts were then excised and
placed into the back of each recipient's right ear (Fig. 1E-H).
Electrocardiography and histology
All recipients had electrocardiographs (ECGs) every
2 days after transplantation until the ECG signal disappeared. When
the ECG signal disappeared the recipient mice and rats were
sacrificed. The transplanted hearts were removed, cut into slices
and fixed in 10% formalin for 24 h at room temperature. Standard
histological techniques were followed and the paraffin embedded
samples were cut into 4-µm-thick sections. Following dewaxing the
sections were stained using a standard hematoxylin and eosin
staining method (hematoxylin for 3 min and eosin for 2–3 min at
room temperature) and observed using a light microscope at
magnification, ×100, ×200 and ×400. Survival time of the
transplanted heart was calculated as the mean number of days
between the day of transplantation and the day when the ECG signal
disappeared. The absence of an electrocardiosignal on days 2 and 4
indicated failure of transplant surgery, and the animal would be
excluded from the group.
Transferred gene expression tests and
mixed lymphocyte culture (MLC) tests
Following the disappearance of the
electrocardiosignal, the thymi of all recipients were removed and
ground with a rubber stick to disperse the cells. The dispersed
cells (5×106-1×107/ml) were blocked with 1%
bovine serum albumin (Sigma-Aldrich; Merck KGaA) at 37°C for 4 h,
incubated with primary anti-H-2-K and anti-H-2-D antibodies (as
above; 1:100 dilution) and normal rabbit serum (Sigma-Aldrich;
Merck KGaA) at 4°C for 30 min. The cells were then washed with PBS
and incubated with fluorescein isothiocyanate-conjugated secondary
goat-anti-mouse antibodies (cat. no. 115-005-003; 1:200 dilution;
Jackson ImmunoResearch, West Grove, PA, USA) at 4°C for 30 min and
then subjected to flow cytometry using a flow cytometer with FlowJo
7.6.1 software (FlowJo LLC.; BD Biosciences, Franklin Lakes, NJ,
USA).
Reverse transcription-PCR analyses were performed in
the three mouse recipient groups as previously performed in the
second PCR step. The CDS primers used in the second PCR step of the
gene preparation phase (Table I)
were also used to examine the expression of the seven transferred
loci in recipients.
The histocompatibility between donors and recipients
was assessed using an MLC kit (Tiangen Biotech Co., Ltd.). The
spleens of the recipients and the donors were removed and ground
with a rubber stick to disperse the cells. They were subsequently
cultured in RPMI 1640 medium supplemented with 10% (v/v)
heat-inactivated fetal bovine serum (both Gibco; Thermo Fisher
Scientific, Inc.) in 5% CO2 at 37°C. Spleen cells
collected from the control, 1-vector and 7-vector mice, and the
control and 1-vector rats were mixed with the spleen cells from the
donors' strain or the spleen cells from a third strain from mice at
the UK Institute of Cancer Research (London, UK; ICR) respectively,
and cultured in RPMI 1640 medium supplemented with 10% (v/v)
heat-inactivated fetal bovine serum in 5% CO2 at 37°C
for 24 h. Cell density was then measured using the MTT method. The
purple formazan crystals were dissolved by dimethyl sulfoxide at a
wavelength of 570 nm.
Statistical analysis
Statistical analysis was performed with SPSS 21.0
(IBM Corp., Armonk, NY, USA). A minimum of three repeats were
performed for each experiment. The data are presented as the mean ±
standard deviation. Differences between multiple groups were
analyzed using one-way analysis of variance followed by the
Newman-Keuls test. Differences between two groups were analyzed
using the paired-samples t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Donor's seven loci of the
H-2d genes were obtained
Electrophoresis was performed following the first
PCR step of nested PCR and revealed a β-actin (internal reference)
band, indicating successful extraction of total RNA and reverse
transcription (Fig. 2A). However, as
expected, no product band was detectable. Electrophoresis following
the second PCR step yielded bands that sequence analysis confirmed
were the seven loci (in accordance with the GenBank accession nos.
NW001030614.1, M18523, M33151, AY452201, AY452202, K00971 and
NT039662.2) of the H-2d genes: H-2Kd,
H-2Dd, H-2Ld, H-2Aαd,
H-2Aβd, H-2Eαd and H-2Eβd
(Fig. 2B and C). The yield obtained
with pfu MasterMix was larger and clearer than that obtained with
Tap Platinum Mastermix, therefore pfu MasterMix was use to retrieve
H-2d genes in subsequent experiments.
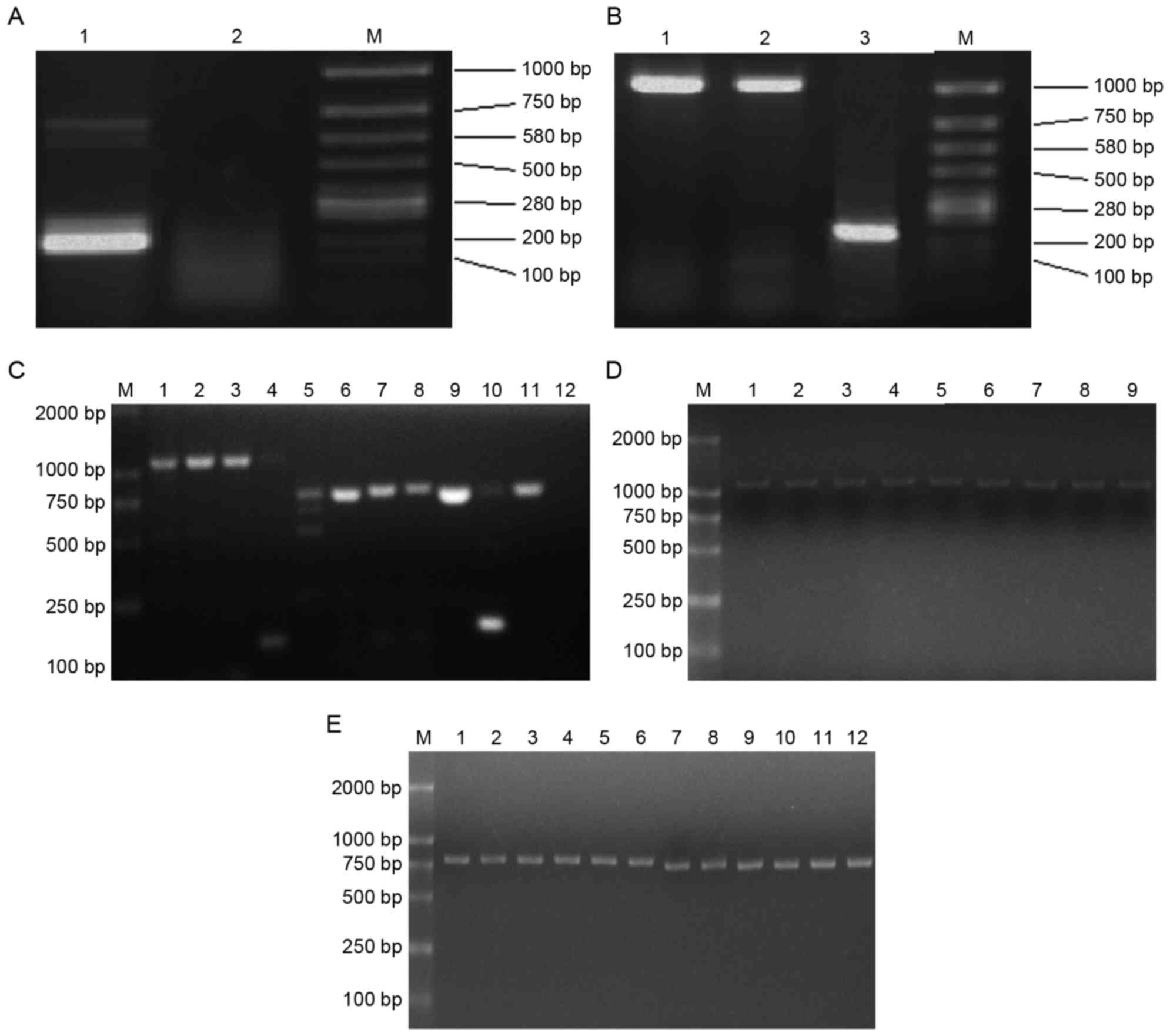 | Figure 2.Donor loci of the H-2d
genes were obtained and expression vectors were constructed. (A)
The first PCR step electro-blottings were indicated. Lane 1,
β-actin; lane 2, the product of PCR; M, marker. (B) The second PCR
step electro-blottings were indicated. Lane 1, the PCR product of
Kd locus by pfu MasterMix; lane 2, product by Tap
Platinum PCR MasterMix; lane 3, β-actin; M, marker. (C) The second
PCR step electro-blottings were indicated. M, marker; lane 1,
Dd locus by pfu MasterMix; lane 2, Dd locus
by Tap Platinum PCR MasterMix; lane 3, Ld locus by pfu
MasterMix; lane 4, Ld locus by Tap Platinum PCR
MasterMix; lane 5, Aαd locus by pfu MasterMix; lane 6,
Aαd locus by Tap Platinum PCR MasterMix; lane 7,
Aβd locus by pfu MasterMix; lane 8, Aβd locus
by Tap Platinum PCR MasterMix; lane 9, Eαd locus by pfu
MasterMix; lane 10, Eαd locus by Tap Platinum PCR
MasterMix; lane 11, Eβd locus by pfu MasterMix; lane 12,
Eβd locus by Tap Platinum PCR MasterMix. (D)
Electro-blottings of PCR results for pCI-neo-H-2d clones
were indicated. M, marker; lanes 1–3, three blottings of
pCI-neo-Kd; lanes 4–6,three blottings of
pCI-neo-Dd; lanes 7–9, three blottings of
pCI-neo-Ld. (E) Electro-blottings of PCR results for
pCI-neo-H-2d clones. M, marker; lanes 1–3, three
blottings of pCI-neo-Aαd; lanes 4–6, three blottings of
pCI-neo-Aβd; lanes 7–9, three blottings of
pCI-neo-Eαd; lanes 10–12, three clones of
pCI-neo-Eβd. PCR, polymerase chain reaction; H-2,
histocompatibility-2 complex. |
Mammalian expression vectors were
constructed
Electrophoresis of PCR results of
pCI-neo-H-2d clones yielded clear bands, demonstrating
that the construction of mammalian expression vector was successful
(Fig. 2D and E). The nested PCR
protocol required two PCR steps; due to this the likelihood of base
mismatches was greater than in simpler protocols. Upon digestion of
pBS-T-H-2ds and ligation of the seven loci of
H-2d genes into pCI-neo to generate
pCI-H-2ds, sequence analysis revealed that the plasmid
in every colony had identical apparent mismatches. Clones
containing base mismatches that had no effect on the amino acid
sequence were selected to transfer donor vectors into the recipient
thymi.
Prolonged ECG signals were indicated
in transgene recipients following heart transplantation
One mouse in the control group succumbed to fatality
during the thymus injection and 1 mouse in the 1-vector group
succumbed during the heart transplantation. A total of 5 mice
succumbed on 1 or 2 days after transplantation (3 in the control
group, 1 in the 1-vector group and 1 in the 7-vector group). No
rats succumbed to fatality during the injection or following
transplant. On days 2 and 4 after transplantation, no
electrocardiosignal was detected in 11-vector mouse, 11-vector rat
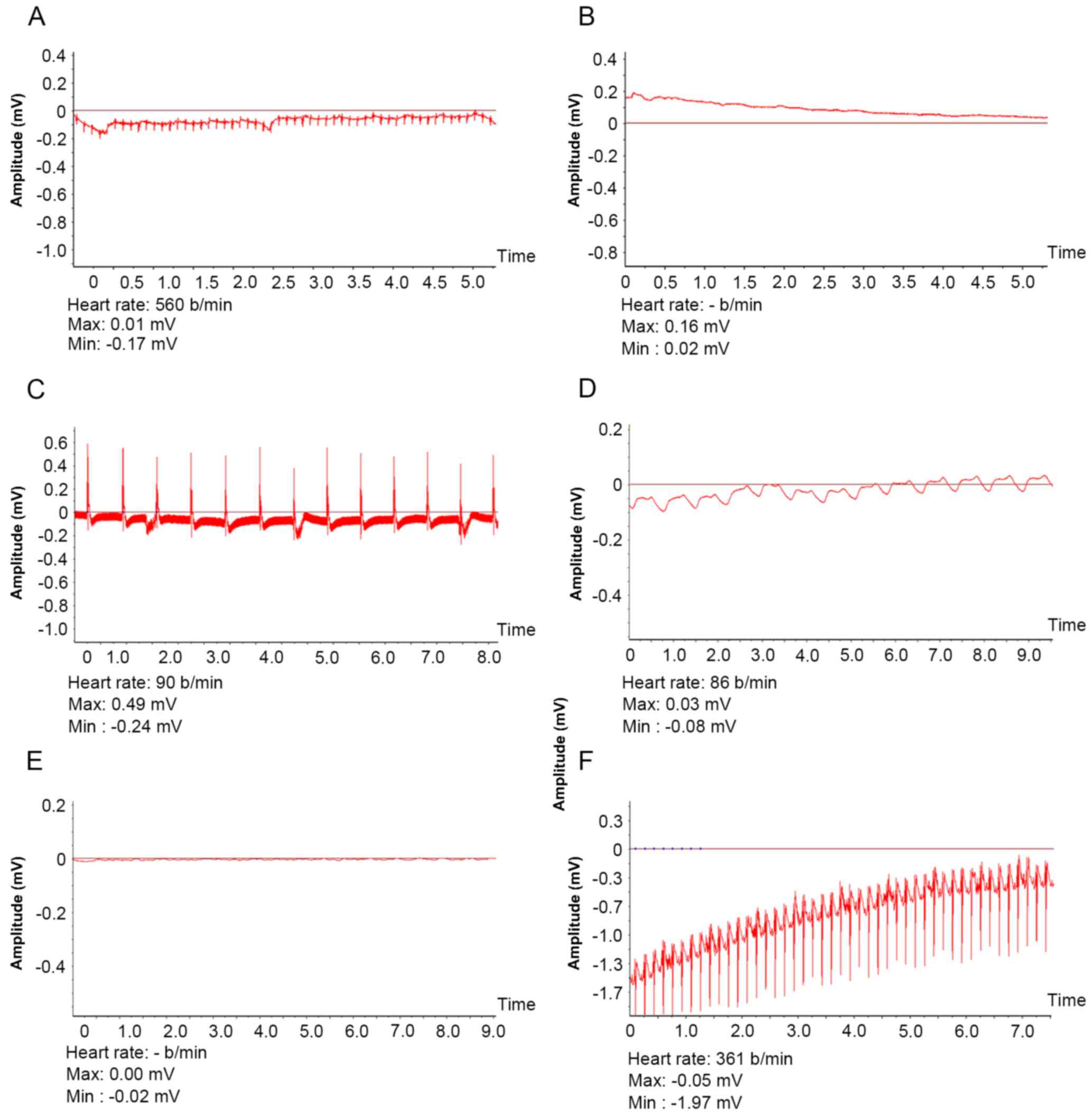
and 1 control mouse. Electrocardiosignals (Fig. 3) were detected for a significantly
longer duration in the 7-vector mouse group (23.59±6.70 days)
compared with the 1-vector (16.67±6.6 days; P<0.05) or control
mouse group (9.11±2.75 days; P<0.01). In rats,
electrocardiosignals were significantly longer in 1-vector rats
compared with the control rate (14.61±2.98 vs. 6.40±1.58 days;
P<0.01).
Expression of donor H-2d
genes in recipient thymi
Following the disappearance of the ECG signal, the
thymi of the recipients were removed and subjected to flow
cytometry and PCR analysis. In the 1-vector mice, the positive
ratio of H-2Kd was 36.83±8.96% and H-2Dd was
negative (Fig. 4A and B). In the
7-vector mice, the positive ratio of H-2Kd was
43.61±6.35% and that of H-2Dd was 50.08±7.21% (Fig. 4C and D). In 1-vector rats, the
positive ratio of H-2Kd was 60.69±1.06% (Fig. 4E). H-2Kd and
H-2Dd were negative in control mice and H-2Kd
was negative in control rats (data not shown). Electrophoresis
following PCR yielded bands of H-2Kd in 1-vector mice
(Fig. 4F) and seven loci of the
H-2d genes in 7-vector mice (Fig. 4G). There was no H-2d band
in control mice (data not shown). Mouse β-actin mRNA bands appeared
in the three mouse groups. Taken together, the flow cytometry and
PCR results indicate that the H-2d genes were
transferred into and expressed in the mouse and rat recipients'
thymus stromal cells.
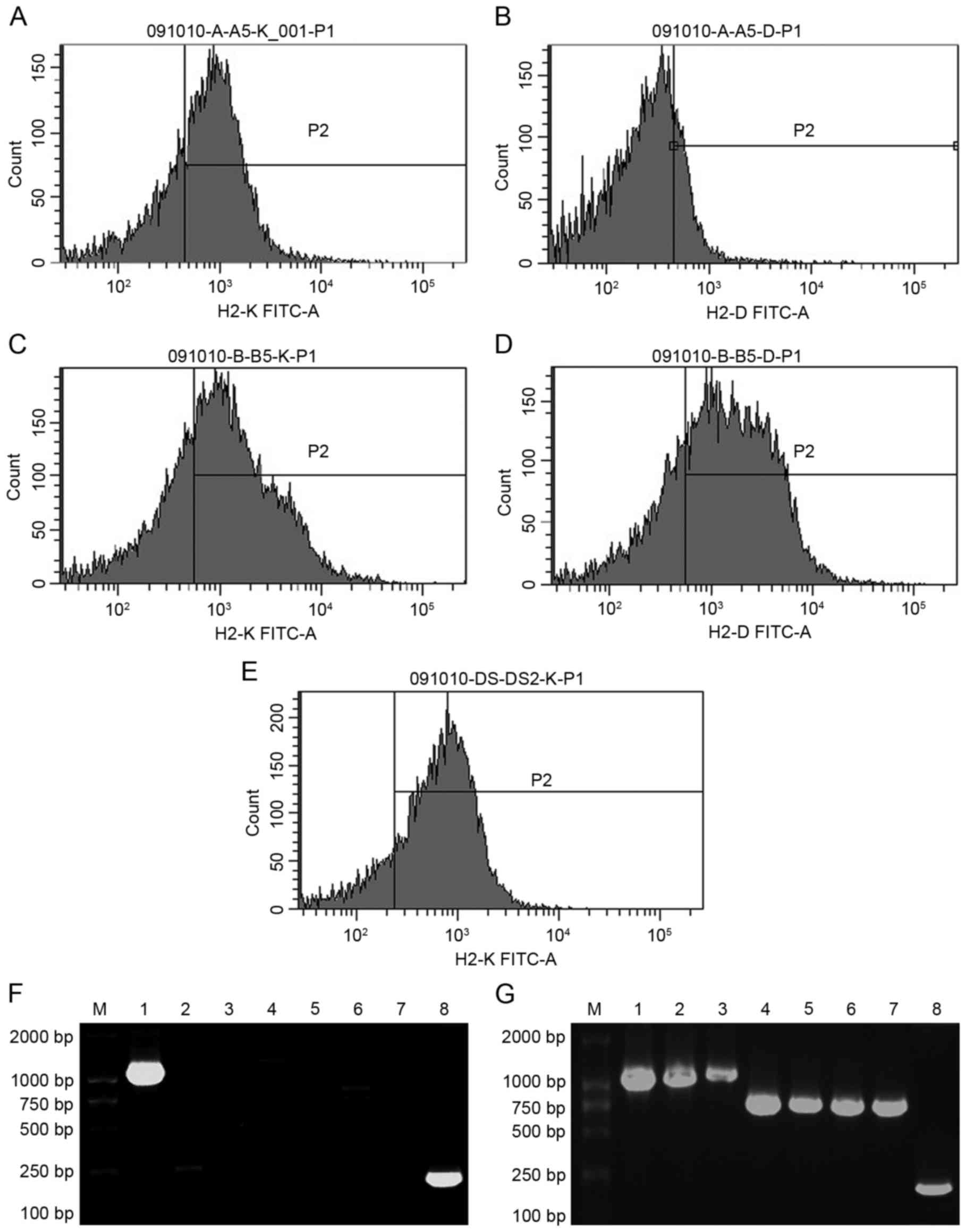 | Figure 4.Donor H-2d genes express
in recipient thymi. Flow cytometry results of the 1-vector mice (A)
incubated with anti-mouse H-2Kd antibodies and (B)
anti-mouse H-2Dd antibodies were determined. Flow
cytometry results of the 7-vector mice (C) incubated with
anti-mouse H-2Kd antibodies and (D) incubated with
anti-mouse H-2Dd antibodies were also indicated. (E)
Flow cytometry results of the experimental (1-vector) rats
incubated with anti-mouse H-2Kd antibodies were
indicated. PCR results of the thymus cells from (F) 1-vector
recipient mice (the bands of H-2Kd and mouse β-actin are
clear) and (G) 7-vector recipient mice (all the bands of seven
H-2d loci and mouse β-actin are clear) were obtained.
PCR, polymerase chain reaction; H-2, histocompatibility-2 complex.
Lane 1, H-2Kd; lane 2, H-2Dd; lane 3,
H-2Ld; lane 4, H-2Aαd; lane 5,
H-2Aβd; lane 6, H-2Eαd; lane 7,
H-2Eβd and lane 8, β-actin. |
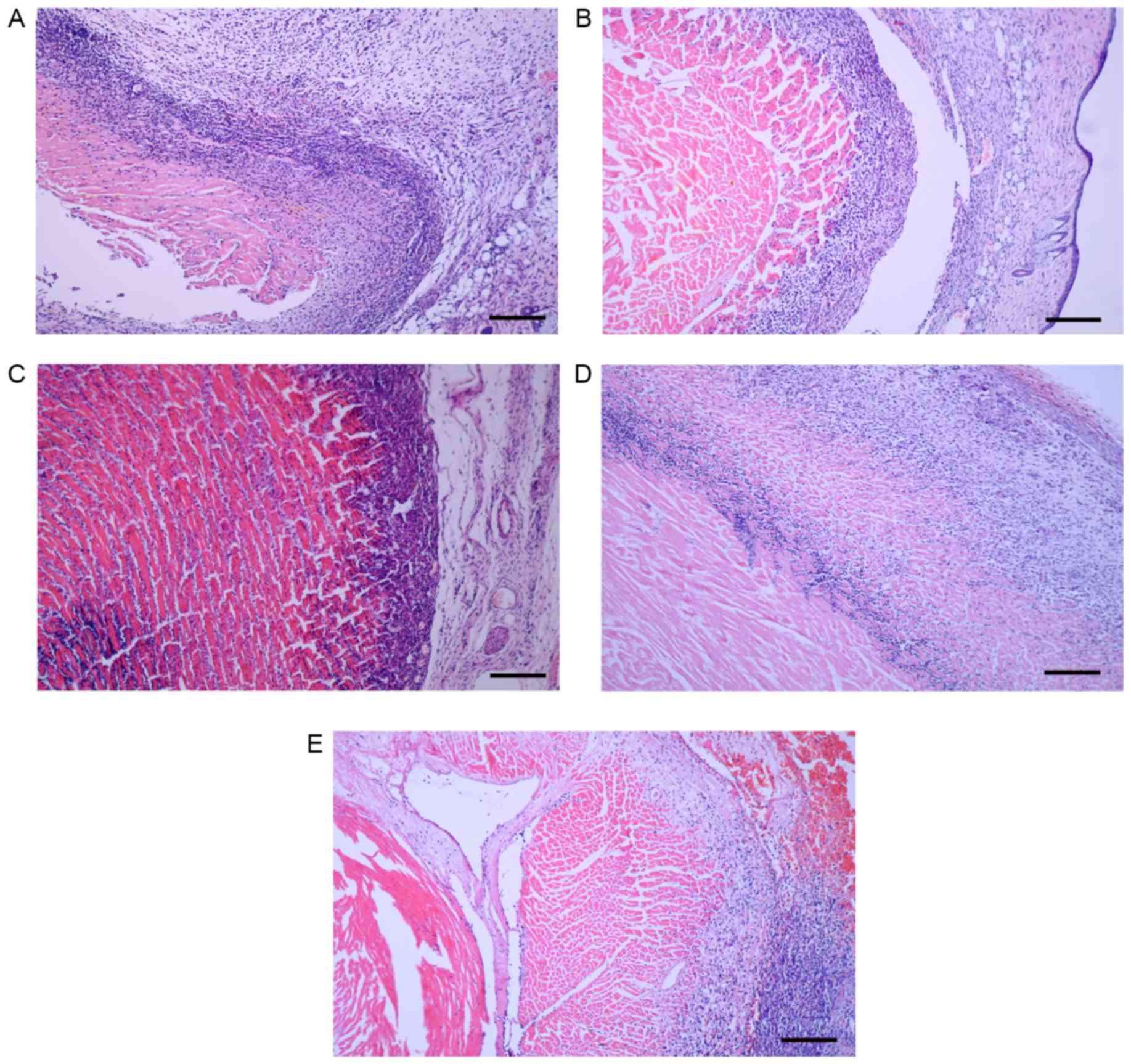
Histological analysis demonstrated
that the transgene recipients were healthy
Histological analysis of sections of transplanted
hearts revealed that the skin around the heart was clearly
edematous in control mice and rats; edema was reduced in the
experimental groups (Fig. 5).
Control hearts demonstrated marked lymphocyte infiltration,
disruption of the characteristic intracellular structure of the
cardiomyocytes and cell necrosis (Fig.
5A and D). By contrast, experimental hearts in the two species
demonstrated minor lymphocyte infiltration and the intracellular
structure of the cardiomyocytes was retained (Fig. 5B, C and E). These characteristics
differed little between the 1-vector mice and the 7-vector mice.
These results indicate that the rejection was reduced in the
transgene recipients compared with the control recipients in mice
and rats.
High histocompatibility in transgene
recipients
The results of MLC tests are summarized in Table II. Mixtures of spleen cells from
donor mice, recipient mice and recipient rats were cultured and
assessed. The cell density was significantly greater among cells in
the control groups compared with the experimental groups
(P<0.01; Table II), indicating
greater histocompatibility in the latter. Cell density was
significantly greater in 1-vector mice compared with 7-vector mice
(P<0.05), indicating higher histocompatibility in the latter.
The density in the cells from the control and experimental groups
did not significantly differ when they were separately mixed with
cells from a third strain (ICR). As expected, cell densities were
significantly higher when the recipient cells from the experimental
groups were mixed with ICR cells (all P<0.01) compared with when
they were mixed with donor cells. These results indicate that the
histocompatibility with donor cells is higher in transgene
recipients compared with control recipients.
 | Table II.Histocompatibility between donors and
recipients. |
Table II.
Histocompatibility between donors and
recipients.
| Group | Mixed spleen cells
from recipients and donors | Mixed spleen cells
from recipients and ICR | Paired-samples T
test results |
|---|
| Control mice
(n=10) | 0.446±0.021 | 0.442±0.016 | t=0.479,
P>0.05 |
| 1-vector mice
(n=12) |
0.383±0.015a | 0.424±0.018 | t=6.062,
P<0.01 |
| 7-vector mice
(n=14) |
0.362±0.021b | 0.421±0.021 | t=7.433,
P<0.01 |
| Control rats
(n=10) | 0.204±0.058 | 0.218±0.033 | t=0.663,
P>0.05 |
| 1-vector rats
(n=9) | 0.102±0.029 | 0.213±0.045 | t=6.220,
P<0.01 |
Discussion
In the present study, to determine whether a donor's
MHC could be introduced into and expressed in a recipient's thymus,
tolerance to transplantation was assessed. It was assumed that
transferring all seven loci of donor's MHC I and II genes would be
more effective than transferring only the K locus. The results of
the present study demonstrated that transplanted hearts survived
significantly longer in mice in which seven loci were transferred
compared with that in mice in which only the K locus was
transferred (23.59±6.70 vs. 16.67±6.6 days; P<0.05). In
addition, transplanted hearts in the two experimental mouse groups
survived significantly longer compared with those in control mice
(9.11±2.75 days; both P<0.01). Furthermore, the heterograft
transplantation effect was observed by transferring the K locus of
the H-2d genes from donor mice into recipient rats and
by transplanting mice hearts into rats. Once again, the
transplanted hearts survived longer in the experimental group
compared with the control rats (P<0.01). This result suggests
that transferring a donor's MHC gene into a recipient may also be
effective in heterografts.
Although prior introduction of a donor's MHC
antigen, cells or tissue into the recipient has been revealed to
delay allograft rejection (20–23), the
finding that intravenous administration of a donor's MHC antigen to
a recipient does not suppress rejection (24) suggests the allograft may ultimately
be rejected. Consistent with that theory, when Gopinathan et
al (25) intravenously injected
dendritic cells from a donor rat into a recipient, the cells did
not home into the lymph nodes, bone marrow or thymus, and so would
not be expected to have a meaningful impact on rejection.
Additionally, Hillebrands et al (26) revealed that introduction of donor
splenic cells into a recipient thymus prior to transplantation
prolonged survival time somewhat; however, they noted that chronic
rejection could not be suppressed. Similarly, Kobayashi et
al (27) reported that injecting
donor cells into a recipient's thymus did not induce tolerance.
Trani et al (28) suggested
that T cell clonal frequency reduction and transplantation
tolerance induced by intrathymic alloantigen inoculation was
incomplete and transient.
Certain studies have demonstrated that transfecting
a donor's MHC into cells from a recipient and then returning the
transfectants to the recipient did mitigate rejection (29,30).
This approach was used in a previous study by our group. One locus
of a donor's MHC genes was introduced into the recipient thymus
stromal cell in vitro and these transfected cells were
subsequently injected back into the thymus of the recipient prior
to transplantation. The results revealed that the survival time of
transplanted hearts prolonged (17).
However, a limitation of this approach would be that it may take a
prolonged period of time to select cell clones that express the
loci of donor MHC. On the other hand, several studies have reported
the beneficial effects of introducing a plasmid harboring one locus
of a donor's MHC into a recipient (29,31,32).
Therefore, the plasmids of seven loci were introduced to the
recipients thymi directly in the present study.
The function of the thymus in immunity and following
transplantation has been well studied (33–37),
although the findings are somewhat conflicting. When thymus tissue
from a donor was embedded in a renal vesicle to form a composite
thymokidney, which was then transplanted into the recipient,
rejection was mitigated (36,37). The
results from Viret et al (38) are most consistent with a model where,
in addition to the thymocyte/stromal cell interaction avidity,
negative selection is largely determined by accessibility to
self-determinants, regardless of their anatomical distribution in
the thymus. The involvement of multiple stromal cell types in
negative selection may assist in minimizing the chances of
autoreactive T cell escape. In that context, the plasmids of seven
loci of donor H-2d genes were introduced into the
recipient's thymus to induce the deletion of T cells that could
react with the donor's H-2 antigen during negative selection.
Typically a 2 µg sample of total RNA is used to
amplify a given target gene, but as levels of H-2 mRNA of these
seven loci were very low, amplification required a larger amount of
total RNA (39) or the amount
produced would be insufficient. This obstacle was overcome by using
two-step nested PCR as amplifying the RNA twice produced an
increased total amount. The results demonstrated that the amount
produced by the first PCR were too small to visualize the
electrophoresis bands. However the amount productions by the second
PCR was sufficient and the electrophoresis bands were observed
clearly.
In the present study, plasmids containing the
donor's MHC were injected into the recipient's thymus. By
visualizing the procedure using ultrasound, the depth and position
of the injection site was precisely determined. The wounds were not
serious; the mortality rate was 0% in rats and 2.2% in mice.
Although thymus injection under ultrasound guidance in animal
experiments has not, to the best of our knowledge, been previously
reported, the experiences during the current study suggests it is
effective and useful.
In previous studies, transplantation was performed
~1–2 weeks after a donor gene, antigen or donor cells were
introduced into the recipient (40–42). The
aim of the present study was to develop a more clinically relevant
technique by introducing the donor gene and transplanting the donor
hearts on the same day. In the present study, negative selection,
which is a process by which T cells that react with self-MHC
antigen are removed through apoptosis, did not happen immediately
and it was hypothesized that this may explain the shorter survival
time of transplanted organs in the current study compared with
other reports (40–42). However, the results of the present
study were positive overall, suggesting that the negative selection
did work. The positive results suggest that introducing the donor's
MHC into the recipient's thymus may be beneficial, as no
immunosuppressants were used in these experiments but the rejection
was mitigated. The MLC tests performed in the present study
demonstrated that the recipient's cells had less reaction with the
donor's cells in experimental groups compared with that in control
groups. These results suggested a better compatibility between the
recipient and donor tissue and less rejection in the experimental
groups.
Overall, the findings suggested that the expression
of the donor gene and the resultant negative selection of T cells
was sufficient. That is, transfer of the donor MHC (H-2) to the
recipient enabled the recipient, at some level, to recognize the
donor's tissue as self, thereby mitigating rejection.
In conclusion, the long existence of the
electrocardiosignal in the experimental groups, histological
analysis and other results of the present study suggest that
rejection of transplanted hearts may be mitigated substantially by
introducing the donor's MHC into the recipient, however further
studies are warranted.
Acknowledgements
The authors would like to thank Dr. Yueping Lv
(preclinical medical lab, Beijing Chao-Yang Hospital, Capital
Medical University, Beijing, China) for his important directions in
PCR. The authors would also like to thank Miss Pamela Derish
(Department of Surgery, UCSF, San Francisco, CA, USA) for editorial
assistance with the current study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TL, WZ, DMJ, SL and LY conceived and designed the
experiments. WZ, TL, XT, JD and QX performed the experiments. HL,
ZX and SH analyzed the data. SL and XT contributed
reagents/materials/analysis tools. TL, LY and WZ wrote the paper.
LY, JD and ZX reviewed and revised the manuscript. DMJ and JD gave
important directions to the study and revised the manuscript.
Ethics approval and consent to
participate
All experiments were conducted in accordance with
University of California, San Francisco Animal Care and Use
requirements, and approved by the Capital University of Medical
Science Institutional Animal Care and Use Committee (Beijing,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bharat A, Kuo E, Steward N, Aloush A,
Hachem R, Trulock EP, Patterson GA, Meyers BF and Mohanakumar T:
Immunological link between primary graft dysfunction and chronic
lung allograft rejection. Ann Thorac Surg. 86:189–195. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Corris PA and Christie JD: Update in
transplantation 2007. Am J Respir Crit Care Med. 177:1062–1067.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pham VV, Stichtenoth DO and Borlak J:
Graft rejection: Pharmacogenetic analysis or drug anamnesis? Br J
Clin Pharmacol. 65:959–960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peeters P, Van Laecke S and Vanholder R:
Acute kidney injury in solid organ transplant recipients. Acta Clin
Belg. 62 Suppl 2:S389–S392. 2007. View Article : Google Scholar
|
|
5
|
Lechler RI, Sykes M, Thomson AW and Turka
LA: Organ transplantation-how much of the promise has been
realized? Nat Med. 11:605–613. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JI, Sonawane SB, Lee MK, Lee SH, Duff
PE, Moore DJ, O'Connor MR, Lian MM, Deng S, Choi Y, et al: Blockade
of GITR-GITRL interaction maintains Treg function to prolong
allograft survival. Eur J Immunol. 40:1369–1374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peugh WN, Superina RA, Wood KJ and Morris
PJ: The role of H-2 and non-H-2 antigens and genes in the rejection
of murine cardiac allografts. Immunogenetics. 23:30–37. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wood KJ: Principles of transplantation
immunologyOxford Textbook of Medicine. Oxford University Press;
Oxford: pp. 1082003
|
|
9
|
Beck S and Trowsdale J: The human major
histocompatability complex: Lessons from the DNA sequence. Annu Rev
Genomics Hum Genet. 1:117–137. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu CL: Major histocompatibility complex
(introduction)Modern Medical Immunology. Shanghai Medical
University Publishing House; Shanghai: pp. 168–169. 1998
|
|
11
|
Parmar S, Del Lima M, Zou Y, Patah PA, Liu
P, Cano P, Rondon G, Pesoa S, de Padua Silva L, Qazilbash MH, et
al: Donor-recipient mismatches in MHC class I chain-related gene A
in unrelated donor transplantation lead to increased incidence of
acute graft-versus-host disease. Blood. 114:2884–2887. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang M and Shen GX: The differentiation of
T cellMedical Immunology. Gong FL: Science Publishing House;
Beijing: pp. 151–153. 2000
|
|
13
|
Maurice D, Hooper J, Lang G and Weston K:
c-Myb regulates lineage choice in developing thymocytes via its
target gene Gata3. EMBO J. 26:3629–3640. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldrath AW and Bevan MJ: Selecting and
maintaining a diverse T-cell repertoire. Nature. 402:255–262. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kishimoto H and Sprent J: The thymus and
central tolerance. Clin Immunol. 95:S3–S7. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kruskall MS: The major histocompatibility
complex: The value of extended haplotypes in the analysis of
associated immune diseases and disorders. Yale J Biol Med.
63:477–486. 1990.PubMed/NCBI
|
|
17
|
Li T, Yan J, Tan JL, Lv YP, Hou SC, Li ST,
Xu Q, Tong XH, Ding J, Zhang Zt and Li H: Donor MHC gene to
mitigate rejection of transplantation in recipient mice. Chin Med J
(Engl). 124:4279–4285. 2011.PubMed/NCBI
|
|
18
|
Judd KP and Trentin JJ: Cardiac
transplantation in mice. Transplantation. 11:298–308.
1971.PubMed/NCBI
|
|
19
|
Babang G, Morris RE, Babang I and Kates
RE: Evaluation of the in vivo dose-response relationship of
immunosuppressive drugs using a mouse heart transplant model:
Application to cyclosporine. J Pharmacol Exp Ther. 244:259–262.
1988.PubMed/NCBI
|
|
20
|
Fiedor P, Jin MX, Hardy MA and Oluwole SF:
Dependence of acquired systemic tolerance to rat islet allografts
induced by intrathymic soluble alloantigens on host responsiveness,
MHC differences, and transient immunosuppression in the high
responder recipient. Transplantation. 63:279–283. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oluwole SF, Jin MX, Chowdhury NC,
Engelstad K, Ohajekwe OA and James T: Induction of peripheral
tolerance by intrathymic inoculation of soluble alloantigens:
Evidence for the role of host antigen-presenting cells and
suppressor cell mechanism. Cell Immunol. 162:33–41. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Otomo N, Motovama K, Yu S, Shimizu Y,
Margenthaler JA, Tu F and Flye MW: Intrathymic alloantigen-mediated
tolerant, completely MHC-mismatched mouse hearts are specifically
rejected by adoptively transferred in vitro-sensitized anti-class I
L(d+)-specific 2C cells. Transplant Proc. 33:159–160. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strober S: Protective conditioning against
GVHD and graft rejection after combined organ and hematopoietic
cell transplantation. Blood Cell Mol Dis. 40:48–54. 2008.
View Article : Google Scholar
|
|
24
|
Arima T, Lehmann M and Flye MW: Induction
of donor specific transplantation tolerance to cardiac allografts
following treatment with nondepleting (RIB 5/2) or depleting
(OX-38) anti-CD4 mAb plus intrathymic or intravenous donor
alloantigen. Transplantation. 63:284–292. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gopinathan R, DePaz HA, Oluwole OO, Ali
AO, Garrovillo M, Engelstad K, Hardy MA and Oluwole SF: Role of
reentry of in vivo alloMHC peptide-activated T cells into the adult
thymus in acquired systemic tolerance. Transplantation.
72:1533–1541. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hillebrands JL, Raué HP, Klatter FA,
Hylkema MN, Platteel I, Hardonk-Wubbena A, Nieuwenhuis P and Rozing
J: Intrathymic immune modulation prevents acute rejection but not
the development of graft arteriosclerosis (chronic rejection).
Transplantation. 71:914–924. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi E, Kamada N, Delriviere L, Lord
R, Goto S, Walker NI, Enosawa S and Miyata M: Migration of donor
cells into the thymus is not essential for induction and
maintenance of systemic tolerance after liver transplantation in
the rat. Immunology. 84:333–336. 1995.PubMed/NCBI
|
|
28
|
Trani J, Moore DJ, Jarrett BP, Markmann
JW, Lee MK, Singer A, Lian MM, Tran B, Caton AJ and Markmann JF:
CD25+ immunoregulatory CD4 T cells mediate acquired central
transplantation tolerance. J Immunol. 170:279–286. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chowdhury NC, Murphy B, Sayegh MH, Hardy
MA and Oluwole SF: Induction of transplant tolerance by intrathymic
inoculation of synthetic MHC class I allopeptides. Transplant Proc.
29:11361997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sonntag KC, Emery DW, Yasumoto A, Haller
G, Germana S, Sablinski T, Shimizu A, Yamada K, Shimada H, Arn S,
et al: Tolerance to solid organ transplants through transfer of MHC
class II genes. J Clin Invest. 107:65–71. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Spriewald BM, Ensminger SM, Jenkins S,
Morris PJ and Wood KJ: Intrathymic delivery of plasmid-encoding
endoplasmic reticulum signal-sequence-deleted MHC class I
alloantigen can induce long-term allograft survival. Transpl Int.
17:458–462. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ando Y, Beck Y, Ichikawa N, Meigata K,
Nomura Y, Nishimura Y, Tomikawa S and Takiguchi M: Induction of
long-term heart graft survival in HLA class I transgenic mice by
intrathymic injection of HLA class I peptides. Transplant Proc.
30:3890–3891. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
del Rio ML, Pabst O, Ramirez P,
Penuelas-Rivas G, Förster R and Rodriguez-Barbosa JI: The thymus is
required for the ability of FTY720 to prolong skin allograft
survival across different histocompatibility MHC barriers. Transpl
Int. 20:895–903. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Siemionow M, Izycki D, Ozer K, Ozmen S and
Klimczak A: Role of thymus in operational tolerance induction in
limb allograft transplant model. Transplantation. 81:1568–1576.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamamoto S, Teranishi K, Kamano C,
Samelson-Jones E, Arakawa H, Nobori S, Okumi M, Houser S, Shimizu
A, Sachs DH and Yamada K: Role of the thymus in transplantation
tolerance in miniature swine: V. Deficiency of the graft-to-thymus
pathway of tolerance induction in recipients of cardiac
transplants. Transplantation. 81:607–613. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamada K, Vagefi PA, Utsugi R, Kitamura H,
Barth RN, LaMattina JC and Sachs DH: Thymic transplantation in
miniature swine: III. Induction of tolerance by transplantation of
composite thymokidneys across fully major histocompatibility
complex-mismatched barriers. Transplantation. 76:530–536. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nobori S, Samelson-Jones E, Shimizu A,
Hisashi Y, Yamamoto S, Kamano C, Teranishi K, Vagefi PA, Nuhn M,
Okumi M, et al: Long-term acceptance of fully allogeneic cardiac
grafts by cotransplantation of vascularized thymus in miniature
swine. Transplantation. 81:26–35. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Viret C, Sant'Angelo DB, He X, Ramaswamy H
and Janeway CA Jr: A Role for accessibility to
self-peptide-self-MHC complexes in intrathymic negative selection.
J Immunol. 166:4429–4437. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pullen JK, Horton RM, Cai ZL and Pease LR:
Structural diversity of the classical H-2 genes: K, D, and L. J
Immunol. 148:953–967. 1992.PubMed/NCBI
|
|
40
|
Chowdhury NC, Jin MX, Hardy MA and Oluwole
SF: Donor-specific unresponsiveness to murine cardiac allografts
induced by intrathymic-soluble alloantigens is dependent on
alternate pathway of antigen presentation. J Surg Res. 59:91–96.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Knechtle SJ, Wang J, Graeb C, Zhai Y, Hong
X, Fechner JH Jr and Geissler EK: Direct MHC class I complementary
DNA transfer to thymus induces donor-specific unresponsiveness,
which involves multiple immunologic mechanisms. J Immunol.
159:152–158. 1997.PubMed/NCBI
|
|
42
|
Geissler EK, Scherer MN and Graeb C:
Soluble donor MHC class I gene transfer to thymus promotes
allograft survival in a high-responder heart transplant model.
Transpl Int. 13 Suppl 1:S452–S455. 2000. View Article : Google Scholar : PubMed/NCBI
|