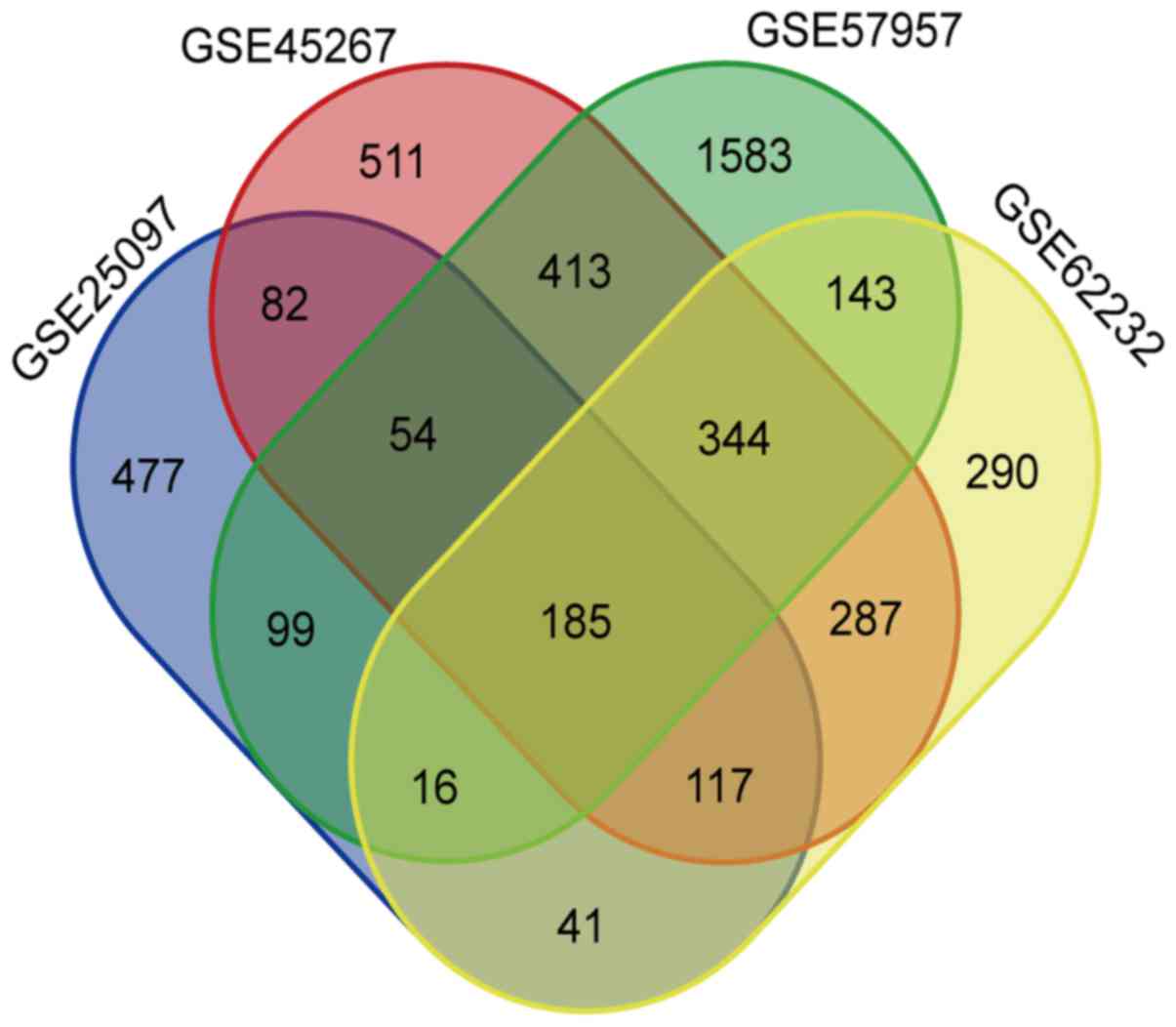
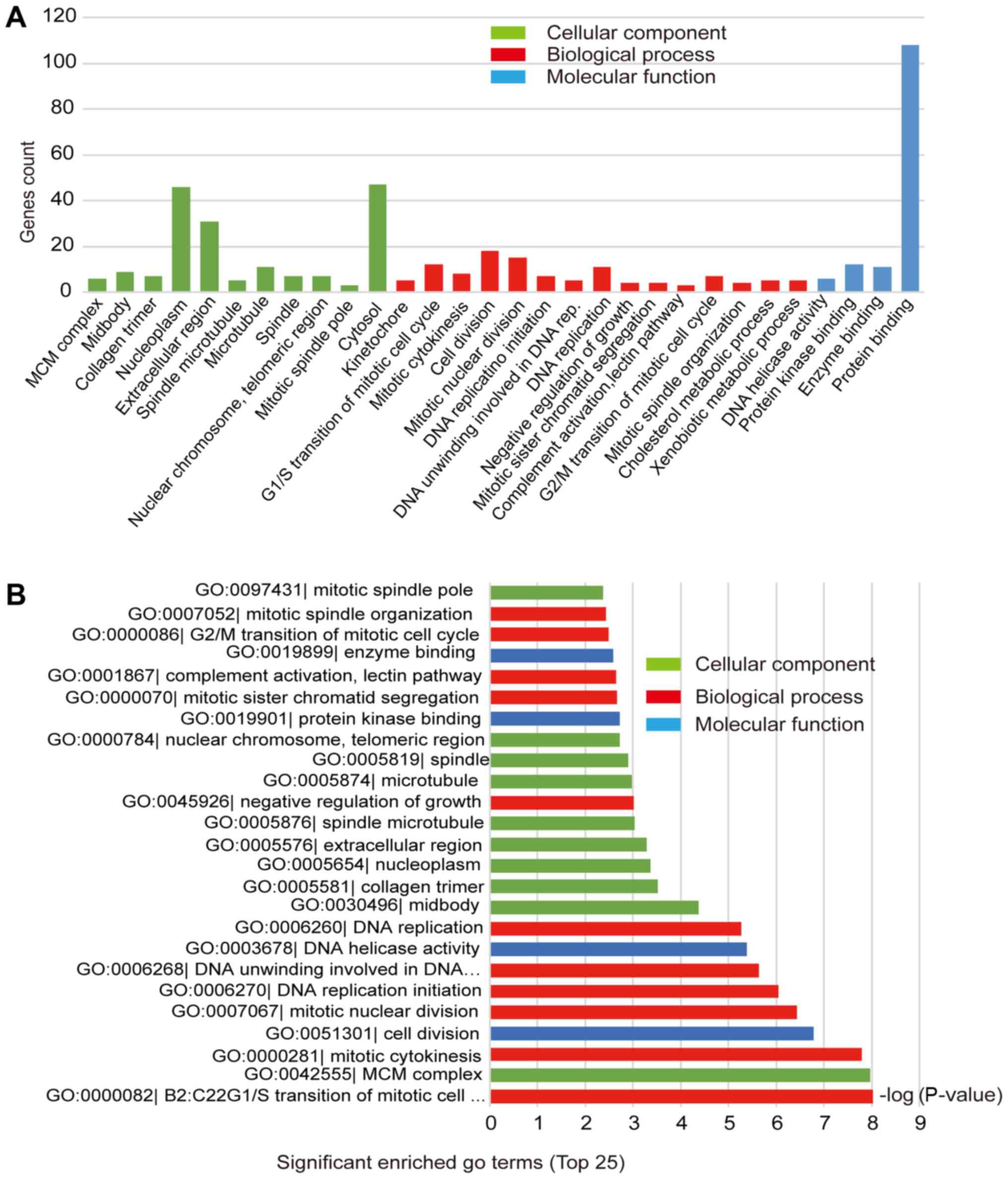
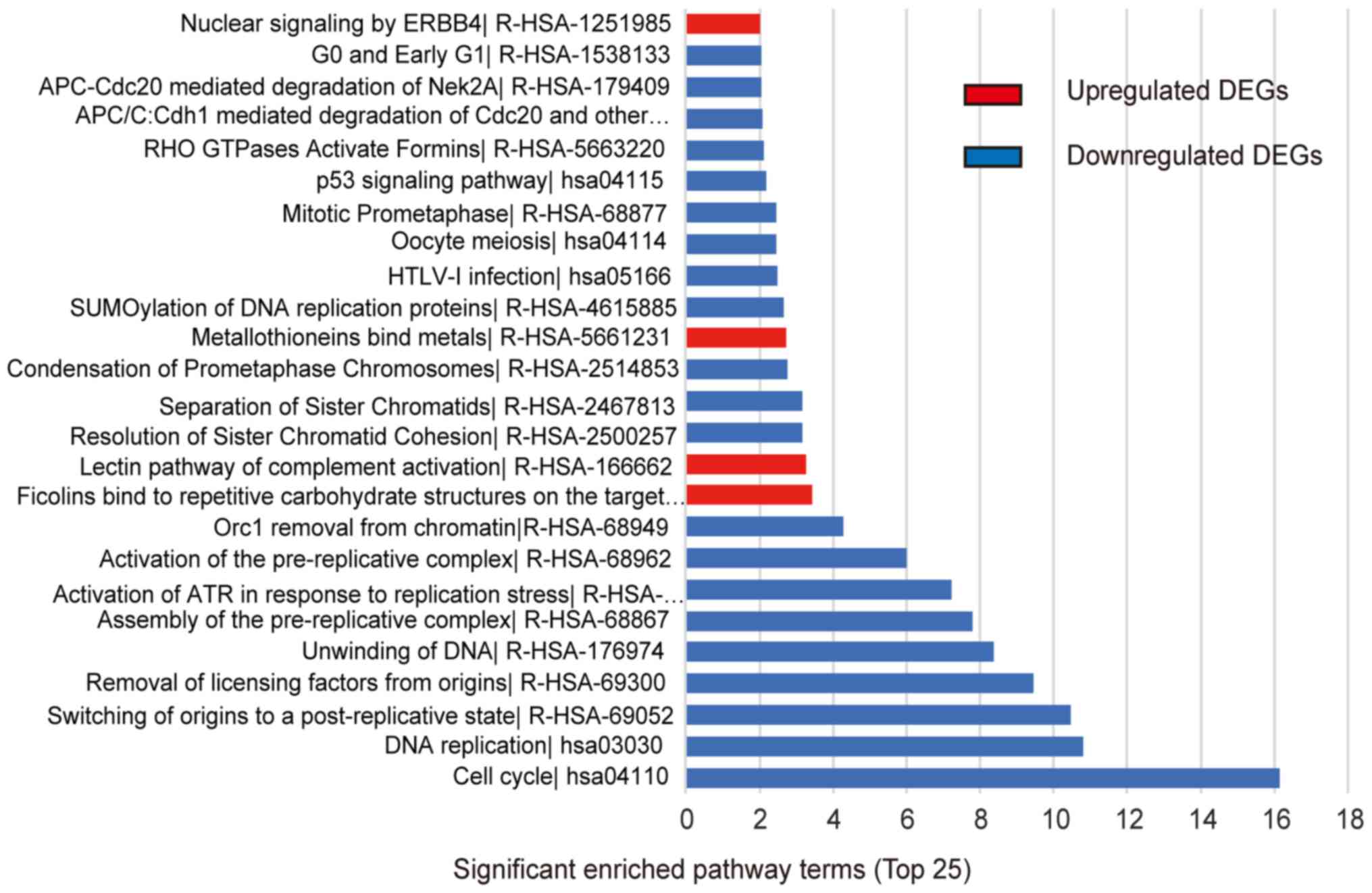
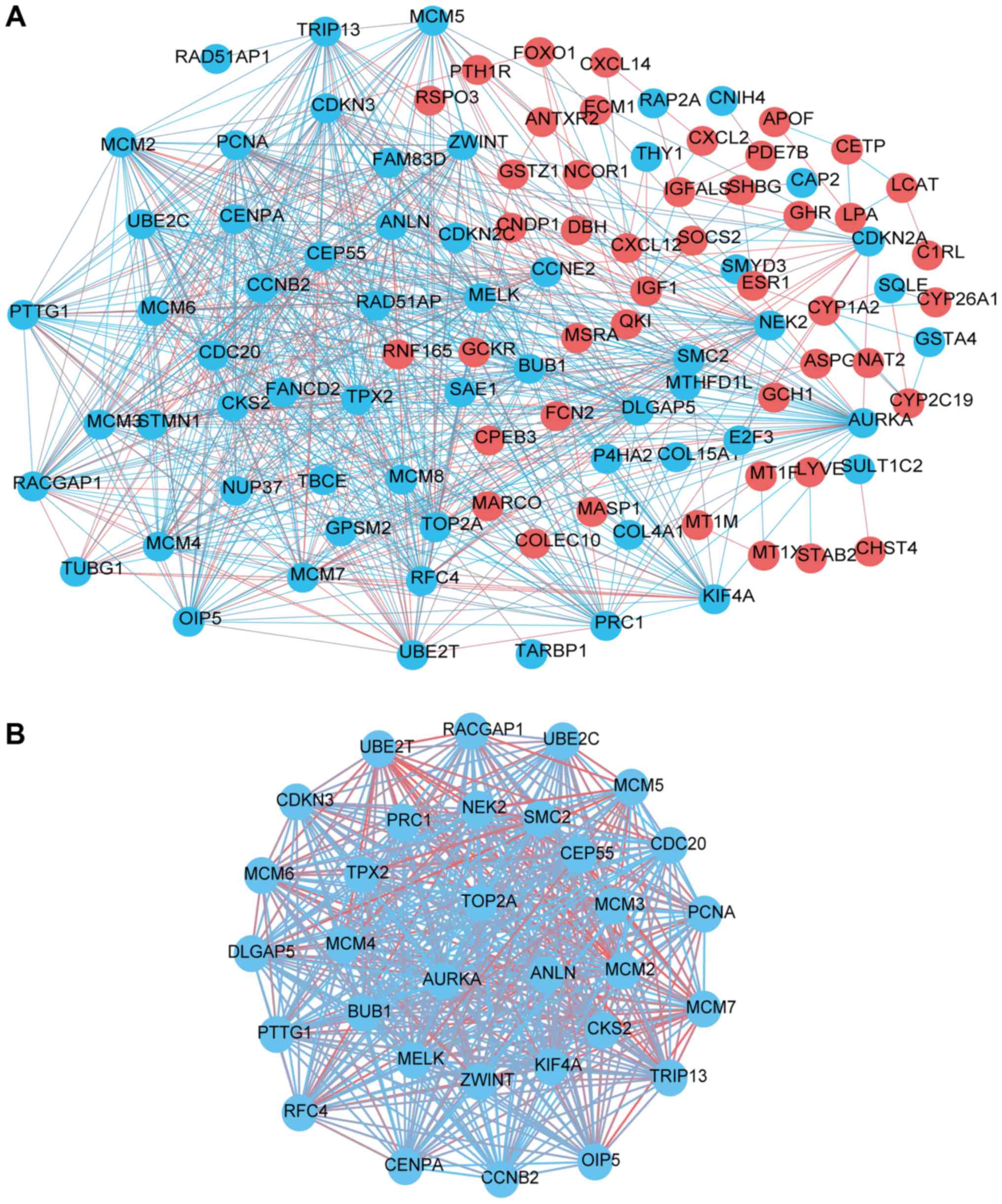
|
1
|
Slotta JE, Kollmar O, Ellenrieder V,
Ghadimi BM and Homayounfar K: Hepatocellular carcinoma: Surgeon's
view on latest findings and future perspectives. World J Hepatol.
7:1168–1183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nair S, Mason A, Eason J, Loss G and
Perrillo RP: Is obesity an independent risk factor for
hepatocellular carcinoma in cirrhosis? Hepatology. 36:150–155.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baffy G, Brunt EM and Caldwell SH:
Hepatocellular carcinoma in non-alcoholic fatty liver disease: An
emerging menace. J Hepatol. 56:1384–1391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valle JW, Dangoor A, Beech J, Sherlock DJ,
Lee SM, Scarffe JH, Swindell R and Ranson M: Treatment of
inoperable hepatocellular carcinoma with pegylated liposomal
doxorubicin (PLD): Results of a phase II study. Br J Cancer.
92:628–630. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Llovet JM and Bruix J: Molecular targeted
therapies in hepatocellular carcinoma. Hepatology. 48:1312–1327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang XW, Hussain SP, Huo TI, Wu CG,
Forgues M, Hofseth LJ, Brechot C and Harris CC: Molecular
pathogenesis of human hepatocellular carcinoma. Toxicology.
181–182:43–47. 2002. View Article : Google Scholar
|
|
10
|
Rinninella E, Zocco MA, De Gaetano A,
Iezzi R, Campanale M, Cesario V, Barbaro F, Ponziani FR, Caracciolo
G, Triarico S, et al: From small nodule to overt HCC: A multistep
process of carcinogenesis as seen during surveillance. Eur Rev Med
Pharmacol Sci. 16:1292–1294. 2012.PubMed/NCBI
|
|
11
|
Brazma A, Parkinson H, Sarkans U,
Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M,
Kemmeren P, Lara GG, et al: ArrayExpress-a public repository for
microarray gene expression data at the EBI. Nucleic Acids Res.
31:68–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schulze K, Imbeaud S, Letouze E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tung EK, Mak CK, Fatima S, Lo RC, Zhao H,
Zhang C, Dai H, Poon RT, Yuen MF, Lai CL, et al:
Clinicopathological and prognostic significance of serum and tissue
Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int.
31:1494–1504. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lebrec JJ, Huizinga TW, Toes RE,
Houwing-Duistermaat JJ and van Houwelingen HC: Integration of gene
ontology pathways with North American Rheumatoid Arthritis
Consortium genome-wide association data via linear modeling. BMC
Proc. 3 Suppl 7:S942009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sung WK, Zheng H, Li S, Chen R, Liu X, Li
Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al: Genome-wide
survey of recurrent HBV integration in hepatocellular carcinoma.
Nat Genet. 44:765–769. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mah WC, Thurnherr T, Chow PK, Chung AY,
Ooi LL, Toh HC, Teh BT, Saunthararajah Y and Lee CG: Methylation
profiles reveal distinct subgroup of hepatocellular carcinoma
patients with poor prognosis. PLoS One. 9:e1041582014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pilati C, Letouze E, Nault JC, Imbeaud S,
Boulai A, Calderaro J, Poussin K, Franconi A, Couchy G, Morcrette
G, et al: Genomic profiling of hepatocellular adenomas reveals
recurrent FRK-activating mutations and the mechanisms of malignant
transformation. Cancer Cell. 25:428–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM,
Sung CO, Baek D, Haq F, Ansari AA, Lee SY, et al: Genomic portrait
of resectable hepatocellular carcinomas: Implications of RB1 and
FGF19 aberrations for patient stratification. Hepatology.
60:1972–1982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ejlertsen B, Tuxen MK, Jakobsen EH, Jensen
MB, Knoop AS, Højris I, Ewertz M, Balslev E, Danø H, Vestlev PM, et
al: Adjuvant cyclophosphamide and docetaxel with or without
epirubicin for early TOP2A-normal breast cancer: DBCG 07-READ, an
open-label, phase III, randomized trial. J Clin Oncol.
35:2639–2646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Di Leo A, Desmedt C, Bartlett JM, Piette
F, Ejlertsen B, Pritchard KI, Larsimont D, Poole C, Isola J, Earl
H, et al: HER2 and TOP2A as predictive markers for
anthracycline-containing chemotherapy regimens as adjuvant
treatment of breast cancer: A meta-analysis of individual patient
data. Lancet Oncol. 12:1134–1142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fritz P, Cabrera CM, Dippon J, Gerteis A,
Simon W, Aulitzky WE and van der Kuip H: c-erbB2 and topoisomerase
IIalpha protein expression independently predict poor survival in
primary human breast cancer: A retrospective study. Breast Cancer
Res. 7:R374–R384. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lazaris AC, Kavantzas NG, Zorzos HS,
Tsavaris NV and Davaris PS: Markers of drug resistance in relapsing
colon cancer. J Cancer Res Clin Oncol. 128:114–118. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Costa MJ, Hansen CL, Holden JA and Guinee
D Jr: Topoisomerase II alpha: Prognostic predictor and cell cycle
marker in surface epithelial neoplasms of the ovary and peritoneum.
Int J Gynecol Pathol. 19:248–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients survival and chemoresistance. Int J Cancer.
124:644–652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stoimenov I and Helleday T: PCNA on the
crossroad of cancer. Biochem Soc Trans. 37:605–613. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoon SM, Gerasimidou D, Kuwahara R,
Hytiroglou P, Yoo JE, Park YN and Theise ND: Epithelial cell
adhesion molecule (EpCAM) marks hepatocytes newly derived from
stem/progenitor cells in humans. Hepatology. 53:964–973. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cai X, Zhai J, Kaplan DE, Zhang Y, Zhou L,
Chen X, Qian G, Zhao Q, Li Y, Gao L, et al: Background progenitor
activation is associated with recurrence after hepatectomy of
combined hepatocellular-cholangiocarcinoma. Hepatology.
56:1804–1816. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Visakorpi T: Proliferative activity
determined by DNA flow cytometry and proliferating cell nuclear
antigen (PCNA) immunohistochemistry as a prognostic factor in
prostatic carcinoma. J Pathol. 168:7–13. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dworakowska D, Gozdz S, Jassem E, Badzio
A, Kobierska G, Urbaniak A, Skokowski J, Damps I and Jassem J:
Prognostic relevance of proliferating cell nuclear antigen and p53
expression in non-small cell lung cancer. Lung Cancer. 35:35–41.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang SC, Nakajima Y, Yu YL, Xia W, Chen
CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R and Hung MC:
Tyrosine phosphorylation controls PCNA function through protein
stability. Nat Cell Biol. 8:1359–1368. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jeng YM, Peng SY, Lin CY and Hsu HC:
Overexpression and amplification of Aurora-A in hepatocellular
carcinoma. Clin Cancer Res. 10:2065–2071. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen SS, Chang PC, Cheng YW, Tang FM and
Lin YS: Suppression of the STK15 oncogenic activity requires a
transactivation-independent p53 function. EMBO J. 21:4491–4499.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dauch D, Rudalska R, Cossa G, Nault JC,
Kang TW, Wuestefeld T, Hohmeyer A, Imbeaud S, Yevsa T, Hoenicke L,
et al: A MYC-aurora kinase A protein complex represents an
actionable drug target in p53-altered liver cancer. Nat Med.
22:744–753. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tokunaga M, Uto H, Oda K, Tokunaga M,
Mawatari S, Kumagai K, Haraguchi K, Oketani M, Ido A, Ohnou N, et
al: Influence of human T-lymphotropic virus type 1 coinfection on
the development of hepatocellular carcinoma in patients with
hepatitis C virus infection. J Gastroenterol. 49:1567–1577. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu B, Hong S, Tang Z, Yu H and Giam CZ:
HTLV-I Tax directly binds the Cdc20-associated anaphase-promoting
complex and activates it ahead of schedule. Proc Natl Acad Sci USA.
102:63–68. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin B, Wang W, Du G, Huang GZ, Han LT,
Tang ZY, Fan DG, Li J and Zhang SZ: Identifying hub genes and
dysregulated pathways in hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 19:592–601. 2015.PubMed/NCBI
|
|
41
|
Calle EE and Kaaks R: Overweight, obesity
and cancer: Epidemiological evidence and proposed mechanisms. Nat
Rev Cancer. 4:579–591. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
White DL, Kanwal F and El-Serag HB:
Association between nonalcoholic fatty liver disease and risk for
hepatocellular cancer, based on systematic review. Clin
Gastroenterol Hepatol. 10:1342–1359.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the IL-6/STAT3 signaling pathway.
Hepatology. 67:171–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sharma SA, Kowgier M, Hansen BE, Brouwer
WP, Maan R, Wong D, Shah H, Khalili K, Yim C, Heathcote EJ, et al:
Toronto HCC risk index: A validated scoring system to predict
10-year risk of HCC in patients with cirrhosis. J hepatol. Aug
24–2017.doi: 10.1016/j.jhep.2017.07.033.
|