|
1
|
Chen J and Liu C: Sulfasalazine for
ankylosing spondylitis. Cochrane Database Syst Rev: CD004800. 2005.
View Article : Google Scholar
|
|
2
|
Bleil J, Maier R, Hempfing A, Schlichting
U, Appel H, Sieper J and Syrbe U: Histomorphologic and
histomorphometric characteristics of zygapophyseal joint remodeling
in ankylosing spondylitis. Arthritis Rheumatol. 66:1745–1754. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sykes M, Doll H and Gaffney K: Comment on:
‘Diagnostic delay in patients with rheumatoid arthritis, psoriatic
arthritis and ankylosing spondylitis: Results from the Danish
nationwide DANBIO registry’ by Sørensen et al. Ann Rheum
Dis. 73:e442014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brown MA, Kenna T and Wordsworth BP:
Genetics of ankylosing spondylitis-insights into pathogenesis. Nat
Rev Rheumatol. 12:81–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Colbert RA, Tran TM and Layh-Schmitt G:
HLA-B27 misfolding and ankylosing spondylitis. Mol Immunol.
57:44–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin TT, Lu J, Qi CY, Yuan L, Li XL, Xia LP
and Shen H: Elevated serum level of IL-27 and VEGF in patients with
ankylosing spondylitis and associate with disease activity. Clin
Exp Med. 15:227–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Corr M: Wnt signaling in ankylosing
spondylitis. Clin Rheumatol. 33:759–762. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nibbe RK, Chowdhury SA, Koyutürk M, Ewing
R and Chance MR: Protein-protein interaction networks and
subnetworks in the biology of disease. Wiley Interdiscip Rev Syst
Biol Med. 3:357–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Y, Jing R, Jiang L, Jiang Y, Kuang Q,
Ye L, Yang L, Li Y and Li M: Combination use of protein-protein
interaction network topological features improves the predictive
scores of deleterious non-synonymous single-nucleotide
polymorphisms. Amino Acids. 46:2025–2035. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mar JC, Matigian NA, Quackenbush J and
Wells CA: Attract: A method for identifying core pathways that
define cellular phenotypes. PLoS One. 6:e254452011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolstad B: affy: Built-in processing
methods. 2013.
|
|
14
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:(Database
Issue). D412–D416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szmidt E and Kacprzyk J: The spearman rank
correlation coefficient between intuitionistic fuzzy setsIEEE
international conference on intelligent systems, is 2010. 7–9–July.
2010, University of Westminster; London: pp. 276–280, 2010.
View Article : Google Scholar
|
|
16
|
Liu G, Wong L and Chua HN: Complex
discovery from weighted PPI networks. Bioinformatics. 25:1891–1897.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Srihari S and Leong HW: A survey of
computational methods for protein complex prediction from protein
interaction networks. J Bioinform Comput Biol. 11:12300022013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomita E, Tanaka A and Takahashi H: The
worst-case time complexity for generating all maximal cliques and
computational experiments. Theoretical Computer Sci. 363:28–42.
2006. View Article : Google Scholar
|
|
19
|
Sriganesh S and Ragan MA: Systematic
tracking of dysregulated modules identifies novel genes in cancer.
Bioinformatics. 29:1553–1561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Srihari S and Ragan MA: Systematic
tracking of dysregulated modules identifies novel genes in cancer.
Bioinformatics. 29:1553–1561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bouchard M, Jousselme AL and Doré PE: A
proof for the positive definiteness of the Jaccard index matrix.
Int J Approximate Reasoning. 54:615–626. 2013. View Article : Google Scholar
|
|
22
|
Gabow HN: An efficient implementation of
Edmonds' algorithm for maximum matching on graphs. J ACM (JACM).
23:221–234. 1976. View Article : Google Scholar
|
|
23
|
Mar J: 2011.attract: Methods to Find the
Gene Expression Modules that Represent the Drivers of Kauffman's
Attractor Landscape. Package version 1.18.0.
|
|
24
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Soc Series B
(Methodological). 10:289–300. 1995.
|
|
25
|
Göhre V and Robatzek S: Breaking the
barriers: Microbial effector molecules subvert plant immunity. Annu
Rev Phytopathol. 46:189–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ein-Dor L, Kela I, Getz G, Givol D and
Domany E: Outcome signature genes in breast cancer: Is there a
unique set? Bioinformatics. 21:171–178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Li S, Hao C, Hong G, Zou J, Zhang
Y, Li P and Guo Z: Extracting a few functionally reproducible
biomarkers to build robust subnetwork-based classifiers for the
diagnosis of cancer. Gene. 526:232–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
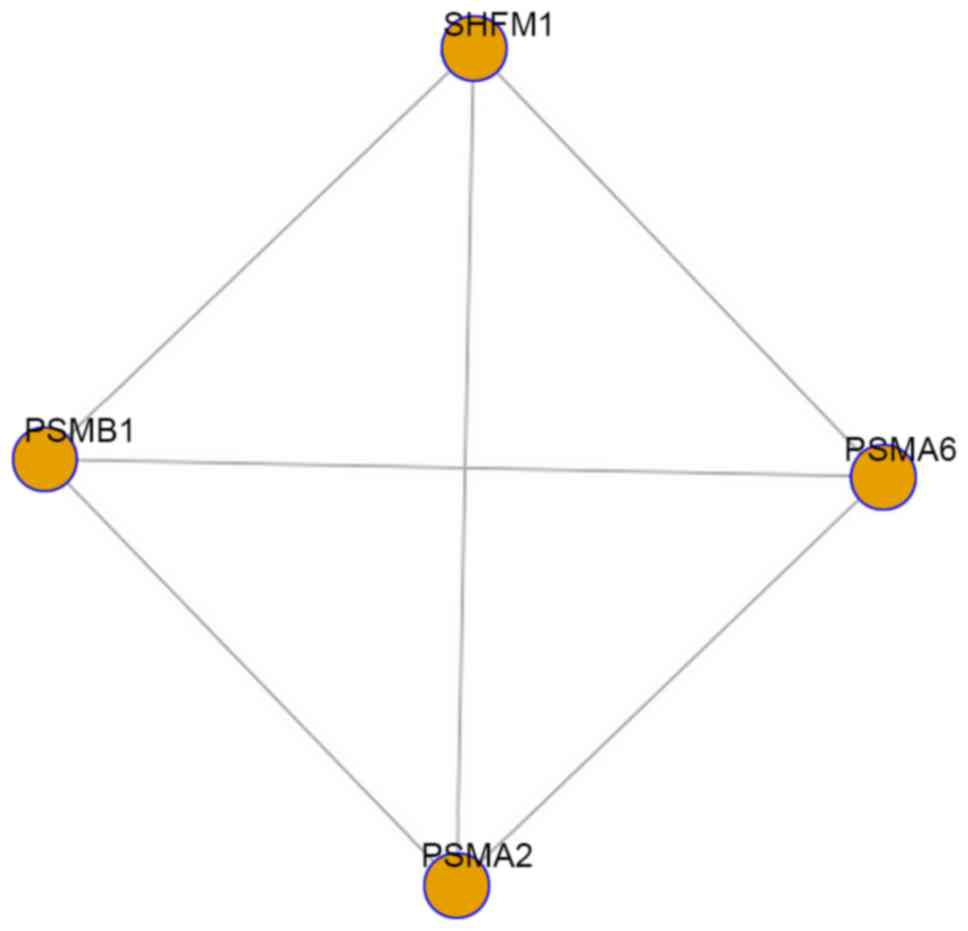
Wu X, Zhao SH, Yu M, Zhu ZM, Wang H, Wang
HL and Li K: Physical mapping of four porcine 20S proteasome core
complex genes (PSMA1, PSMA2, PSMA3 and PSMA6). Cytogenet Genome
Res. 108:3632005. View Article : Google Scholar
|
|
29
|
Wright C, Edelmann M, diGleria K,
Kollnberger S, Kramer H, McGowan S, McHugh K, Taylor S, Kessler B
and Bowness P: Ankylosing spondylitis monocytes show upregulation
of proteins involved in inflammation and the ubiquitin proteasome
pathway. Ann Rheum Dis. 68:1626–1632. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang GC, Zhao MJ and Wu DH: The expression
of proteasome gene in the patients with ankylosing spondylitis.
Chin J Rheumatol. 2003.(In Chinese).
|
|
31
|
Kireva T, Polzer K, Neubert K, Meister S,
Frey B, Baum W, Distler JH, Schett G, Voll RE and Zwerina J:
Proteasome inhibition aggravates tumour necrosis factor-mediated
bone resorption. Annals Rheumatic Dis. 69:61–75. 2010. View Article : Google Scholar
|
|
32
|
Zhao H, Wang D, Fu D and Xue L: Predicting
the potential ankylosing spondylitis-related genes utilizing
bioinformatics approaches. Rheumatol Int. 35:973–979. 2015.
View Article : Google Scholar : PubMed/NCBI
|