Introduction
Endothelial cells are an important feature of blood
vessels, and dysfunction of vascular endothelial cell proliferation
and migration may result in the development of various vascular
diseases, including atherosclerosis, as well as neointimal
proliferation associated with vein graft failure and in-stent
restenosis. Therefore, the proliferation and migration of vascular
endothelial cells serves a key function in vascular diseases, and
an understanding of the associated underlying mechanisms is of
critical importance.
Long non-coding RNAs (lncRNAs) are defined as
transcripts of >200 nucleotides in length that do not encode for
proteins. Initially, lncRNAs were considered to be a transcription
‘noise’, and to not have biological function (1). However, numerous studies have suggested
that lncRNAs serve key functions in the regulation of cell
development, differentiation, proliferation, migration and
apoptosis, as well as other endothelial cell functions and the
pathogenesis of cardiovascular diseases (2–7). At
present, a number of studies have indicated that certain lncRNAs
are involved in the development of cardiovascular disease via the
regulation of endothelial cell proliferation, migration and
apoptosis. For example, Michalik et al (8) demonstrated that lncRNA
metastasis-associated lung adenocarcinoma transcript-1
(lncRNAMALAT1) is involved in the regulation of endothelial cell
function and vascular growth. In diabetic rats, the downregulation
of lncRNAMALAT1 has been revealed to inhibit cardiac myocyte
apoptosis and attenuate left ventricular function (9), as well as suppress the proliferation
and migration of retinal endothelial cells, and attenuate retinal
vascular injury inflammation and function (10). Tao et al (11) demonstrated that downregulation of
lncRNAH19 inhibits the proliferation of cardiac fibroblasts. Pan
(12) revealed that lncRNAH19
regulates the proliferation and apoptosis of human umbilical vein
endothelial cells (HUVECs) and vascular smooth muscle cells (VSMCs)
via modulation of the mitogen-activated protein kinase and nuclear
factor-κB signaling pathways, which subsequently regulate
atherosclerosis formation. Ballantyne et al (13) demonstrated that smooth muscle
enriched lncRNA is involved in regulating the proliferation of
VSMCs and is highly expressed in atherosclerotic plaques.
Furthermore, Qiu et al (14)
revealed that silencing the expression of lncRNA maternally
expressed gene 3 (lncRNAMEG3) promotes endothelial cell
proliferation and angiogenesis; and upregulation of the expression
of lncRNAMEG3 suppresses angiogenesis and the cell cycle in
endothelial cells. These results indicate that lncRNAs serve key
functions in the regulation of endothelial cell function.
Taurine upregulated 1 (TUG1), a 7.1-kb lncRNA, was
first identified in the retinal cells of newborn mice and has been
demonstrated to serve a key function in retinal development
(15). Previous studies have
revealed that lncRNATUG1 is involved in the development of tumors
via regulation of tumor cell proliferation and migration.
Furthermore, TUG1 is highly expressed in vascular endothelial cells
(16,17). Yin et al (6) demonstrated that lncRNATUG1 is highly
expressed in the mouse pancreas and downregulates the expression of
TUG1, which subsequently affects the apoptosis and insulin
secretion of pancreatic β cells, thus suggesting that lncRNATUG1
may represent a new target for the treatment of diabetes. However,
little is known about the association between TUG1 and vascular
diseases. Therefore, investigation into the role of TUG1 is
important with regards to vascular endothelial cells and vascular
diseases.
Rapamycin is a potent immunosuppressive agent, which
is able to inhibit the proliferation and migration of endothelial
cells and angiogenesis. Rosner et al (18) identified that rapamycin inhibits
human in-stent restenosis by inhibiting the proliferation and
migration of vascular smooth muscle cells. Kawatsu et al
(19) identified that rapamycin
inhibits autologous vein graft restenosis by impeding venous
neointimal hyperplasia. Although treatment with rapamycin reduces
the incidence of postoperative restenosis by inhibiting the
proliferation and migration of vascular smooth muscle cells, it
increases the incidence of thrombosis. However, although certain
drugs may be used to treat the occurrence of thrombus, they may be
non-responsive and exhibit an efficacy that is below expectations
(20–23). To date, the mechanism by which
rapamycin inhibits the proliferation and migration of endothelial
cells has been studied (24);
however, the underlying mechanisms remain unclear. Recent research
has revealed that lncRNAs serve key functions in the regulation of
cell proliferation and migration. Li et al (25) reported that the lncRNA HOX transcript
antisense intergenic RNA regulates the AKT/mammalian target of
rapamycin (mTOR) signaling pathway, which subsequently enhances
osteosarcoma cell proliferation and metastasis. Wang et al
(26) demonstrated that lncRNA
colorectal neoplasia differentially expressed promotes the
proliferation and invasion of glioma cells by regulating the mTOR
pathway. Matsumoto et al (27) demonstrated that the polypeptide
encoded by LINC00961 serves a key function in regulating mTOR
complex 1 activity. These results indicate that lncRNAs may be
involved in regulating the function of endothelial cells treated
with rapamycin. Therefore, it is important to determine the
underlying molecular mechanism associated with inhibition of the
proliferation and migration of rapamycin-treated vascular
endothelial cells. The current study aimed to investigate the
effect of rapamycin on the proliferation, migration and apoptosis
of endothelial cells, as well as to investigate the association of
lncRNATUG1 with these processes.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs; cat.
no. CRL-1730) were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). HUVECs were cultured in
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) and 10 ng/ml vascular
endothelial growth factor (VEGF; PeproTech, Inc., Rocky Hill, NJ,
USA) at 37°C in 5% CO2. All experiments were performed
according to the manufacturer's protocol.
Small-interfering RNA (siRNA) and cell
transfection
siRNA specifically targeting TUG1 (siTUG1: siTUG1-1,
5′-GCUUGGCUUCUAUUCUGAAUCCUUU-3′; siTUG1-2,
5′-CAGCUGUUACCAUUCAACUUCUUAA-3′) and negative control siRNA (siNC,
5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Once HUVECs reached 50–70%
confluence, they were seeded in 6-well plates and transfected with
100 nM siTUG1 and siNC using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 24 h post-transfection, DMEM and 100
ng/ml rapamycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
were added. Subsequently, the cells were harvested for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analysis.
RT-qPCR
Total RNA was extracted from cells using RNAiso Plus
reagent (Takara Biotechnology Co., Ltd., Dalian, China) according
to the manufacturer's protocol. cDNA was synthesized from the
extracted RNA using the Fast Quant RT kit (Tiangen Biotech Co.,
Ltd., Beijing, China) according to the manufacturer's protocol.
qPCR was performed using a Super Real PreMix Plus kit (Tiangen
Biotech Co., Ltd.) and the ABI 7500 qPCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions used for qPCR were as follows: 95°C for 5 min, followed
by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The sequences
of primers used for qPCR were as follows: TUG1 forward,
5′-TCCAGACCCTCAGTGCAAAC-3′ and reverse, 5′-TGTTGTGGTGTATGTGGGCA-3′;
VEGF forward, 5′-CTACCTCCACCATGCCAAGT-3′ and reverse,
5′-GCAGTAGCTGCGCTGATAGA-3′; matrix metalloproteinase (MMP)-2
forward, 5′-TGATGGCATCGCTCAGATCC-3′ and reverse,
5′-GGCCTCGTATACCGCATCAA-3′; MMP-9 forward,
5′-GTCATCCAGTTTGGTGTCGC-3′ and reverse, 5′-GGACCACAACTCGTCATCGT-3′;
B-cell lymphoma 2 (Bcl-2) forward, 5′-TGTGTGTGGAGAGCGTCAAC-3′ and
reverse, 5′-GGGCCGTACAGTTCCACAAA-3′; Caspase3 forward,
5′-ATGGAAGCGAATCAATGGAC-3′ and reverse, 5′-GCTGCATCGACATCTGTACC-3′;
and GAPDH forward, 5′-TCTCTGCTCCTCCTGTTCGA-3′ and reverse,
5′-GCGCCCAATACGACCAAATC-3′. GAPDH was used as an internal control.
The relative gene expression levels were determined using the
2−ΔΔCq method (28).
Cell proliferation assay
A total of ~1×105 HUVECs were seeded into
96-well plates and subsequently incubated for 24 h at 37°C in 5%
CO2. Once the cells reached a confluence of 70%, they
were transfected with siTUG1-1 and siNC using Lipofectamine™ 2000,
according to the manufacturer's protocol. At 24 h
post-transfection, DMEM with or without 100 ng/ml rapamycin was
added to the cells and subsequently 10 µl of CCK-8 reagent (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was added to each
well at 24, 48 and 72 h time intervals. The cells were then
incubated at 37°C for 4 h. The absorbance was measured at 450 nm
with an EnVision® Multi-Mode Plate Reader (PerkinElmer,
Inc., Waltham, MA, USA).
Cell apoptosis analysis
Cell apoptosis was determined using an Annexin
V-FITC/PI Apoptosis kit [MultiSciences (Lianke) Biotech Co., Ltd,
Hangzhou, China]. Then, the cells were harvested by trypsinization
without EDTA via centrifugation at 300 × g for 5 min at 4°C. Cells
(1×106 cells/ml) were then washed twice with cold PBS
and subsequently incubated with 500 µl binding buffer containing 5
µl Annexin V-FITC and 10 µl PI staining solution for 15 min at room
temperature in the dark. Apoptosis was then determined using a BD
FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Results were analyzed using FlowJo 7.6.5 software (FlowJo, LLC,
Ashland, OR USA).
Wound healing assay to evaluate cell
migration
When cells reached a confluence of 90%, 24 h
post-transfection, the monolayer of cells was scraped from each of
the six wells using a sterile 200 µl pipette tip across the
diameter of the wells to form an artificial wound, and the plates
were subsequently washed twice with PBS. Following this, serum-free
medium with/without 100 ng/ml rapamycin was added to the cells,
which were then cultured at 37°C and 5% CO2 for 24 h. An
IX51-A21PH inverted microscope (Olympus Corporation, Tokyo, Japan)
was used to capture images.
Western blot analysis
Total protein was extracted from the cells using
RIPA lysis buffer [MultiSciences (Lianke) Biotech Co., Ltd.]
supplemented with 1 nM PMSF. Following lysis for 30 min, total
protein was isolated via centrifugation at 13,201 × g for 20 min at
4°C. Protein content was then determined using a BCA Protein Assay
kit (Beyotime Institute of Biotechnology, Shanghai, China). A total
of ~20 µg of protein was separated by 10% SDS-PAGE and subsequently
transferred onto polyvinylidene difluoride membranes. Membranes
were blocked using 5% skimmed milk for 2 h at room temperature. The
membranes were then incubated with anti-VEGF (cat. no. ab46154;
1:2,000; Abcam, Cambridge, UK), anti-MMP2 (cat. no. ab37150;
1:2,000; Abcam); anti-MMP9 (cat. no. 3852), anti-Bcl-2 (cat. no.
2870), anti-Caspase3 (cat. no. 9662) and anti-GAPDH (cat. no. 5174;
all 1:1,000; all from Cell Signaling Technology, Inc., Danvers, MA,
USA) overnight at 4°C. Following this, membranes were incubated
with a horseradish peroxidase-labeled secondary antibody (cat. no.
7074; 1:5,000; Cell Signaling Technology, Inc.) for 2 h at room
temperature. Finally, protein bands were detected using an enhanced
chemiluminescence kit (EMD Millipore, Billerica, MA, USA) and a
western blotting detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). GAPDH was used as the reference protein.
Relative protein content was determined using ImageJ software
version 1.48v (National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Data are presented as the mean ± standard deviation. Each
experiment was repeated ≥3 times and differences between two groups
were statistically analyzed using a Student's t-test. Differences
among groups were statistically analyzed using one-way analysis of
variance followed by a Tukey's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
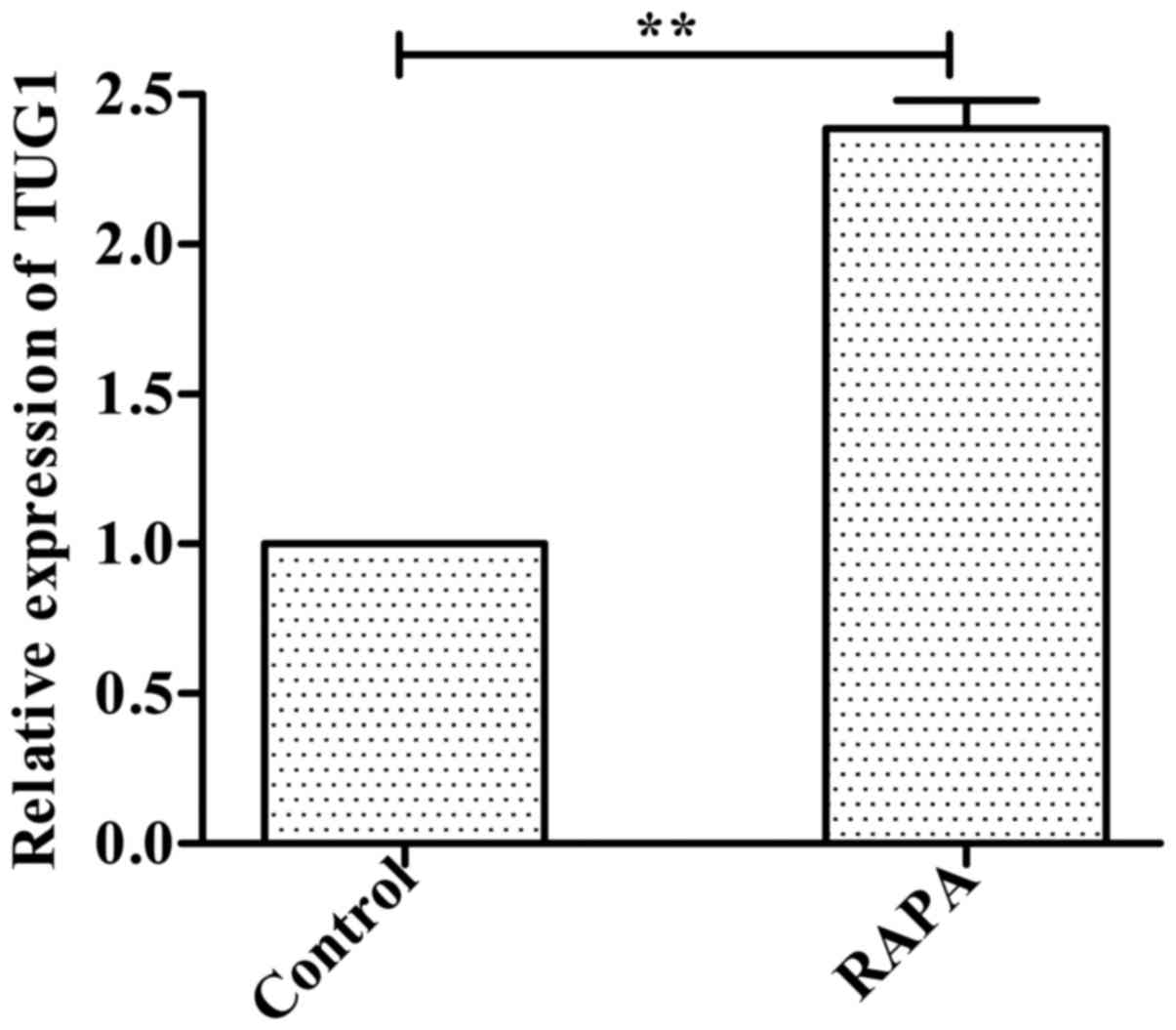
Expression of lncRNATUG1 is
upregulated in HUVECs incubated with rapamycin
Following incubation of HUVECs with 100 ng/ml
rapamycin for 24 h, the RT-qPCR results indicated that expression
of lncRNATUG1 was significantly upregulated compared with the
control group (P<0.01; Fig.
1).
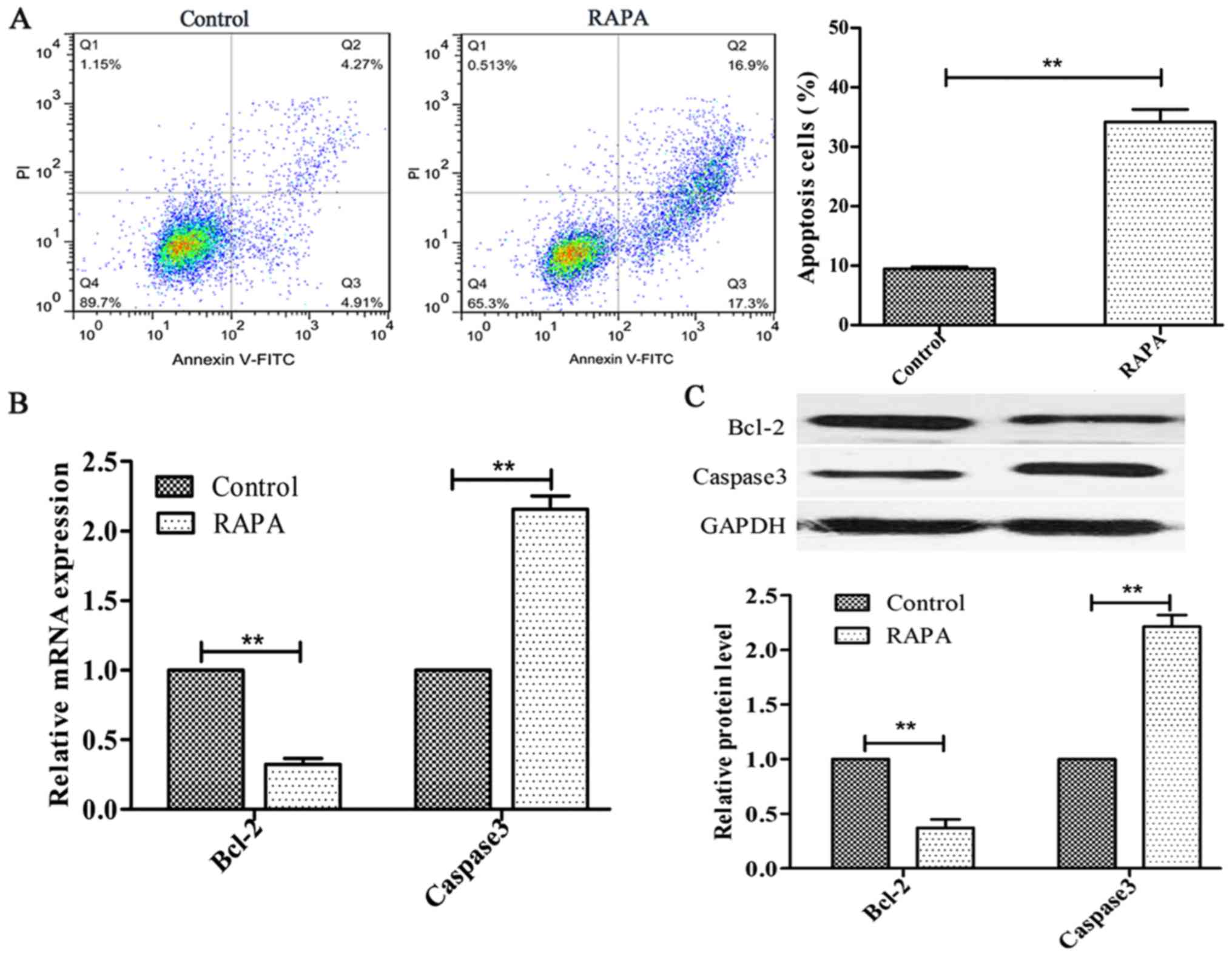
Rapamycin induces cell apoptosis in
HUVECs
In order to investigate the effect of rapamycin on
cell apoptosis, HUVECs were treated with 100 ng/ml rapamycin for 24
h. The results indicated that rapamycin significantly increased
HUVEC apoptosis (P<0.01; Fig.
2A). Additionally, Bcl-2 expression was decreased, whereas the
expression of Caspase3 was increased, following rapamycin treatment
(P<0.01; Fig. 2B and C).
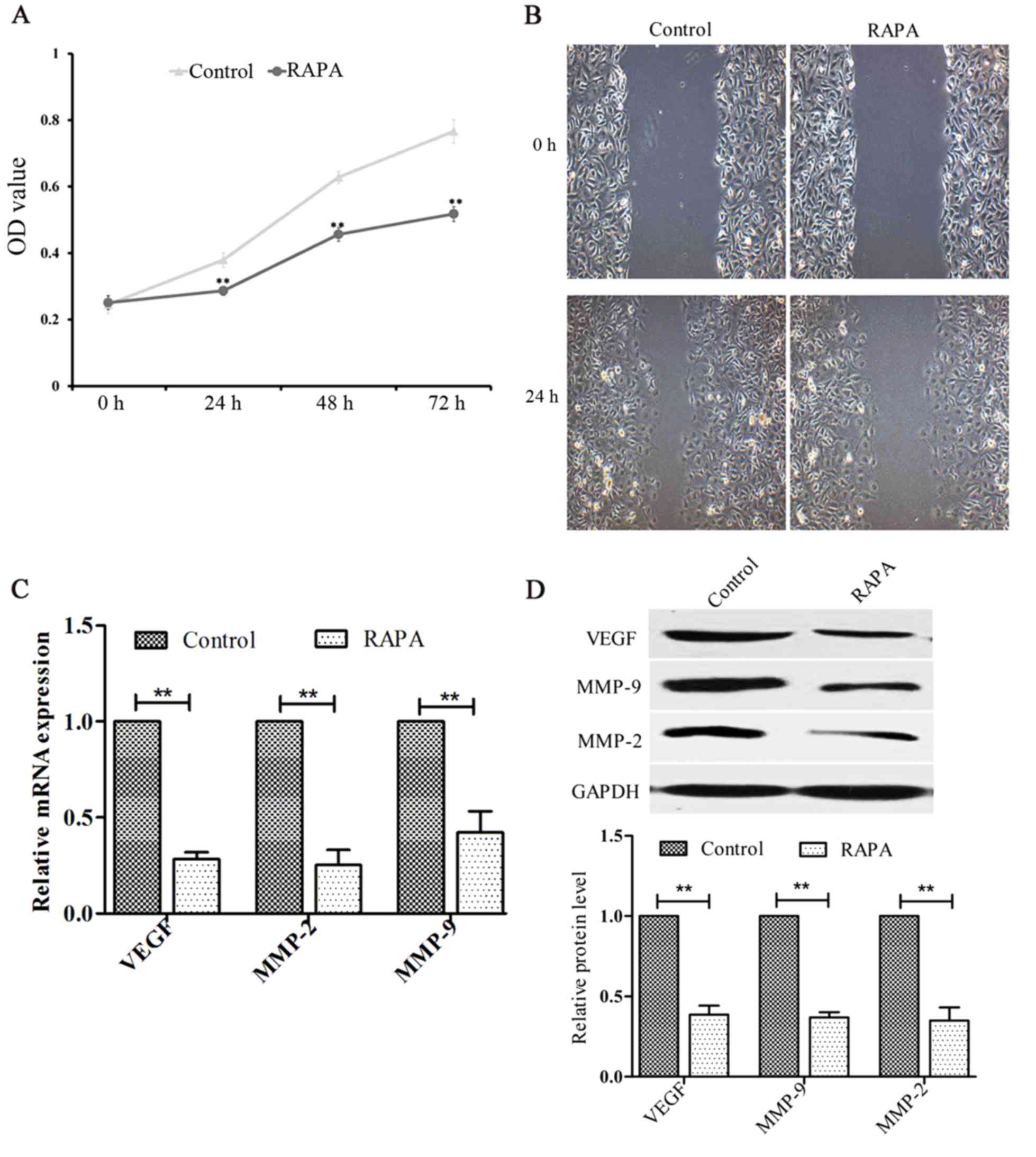
Rapamycin inhibits cell proliferation
and migration in HUVECs
The effects of rapamycin on the proliferation and
migration of HUVECs were investigated using CCK-8 and cell scratch
assays, respectively. The results indicated that rapamycin
inhibited cell proliferation (P<0.01; Fig. 3A) and migration (Fig. 3B). In addition, the levels of VEGF,
MMP-2 and MMP-9 were significantly decreased following treatment
with rapamycin (P<0.01; Fig. 3C and
D).
Expression levels of lncRNATUG1 in
HUVECs are decreased following transfection
In order to downregulate the expression of
lncRNATUG1, lncRNATUG1 siRNAs were transfected into HUVECs. The
results of RT-qPCR analysis indicated that siTUG1-1 and siTUG1-2
significantly inhibited the expression of lncRNATUG1 in HUVECs
(P<0.01; Fig. 4A); however,
siTUG1-1 demonstrated a greater knockdown efficiency compared with
siTUG1-2. Therefore, siTUG1-1 was selected for use in subsequent
experiments.
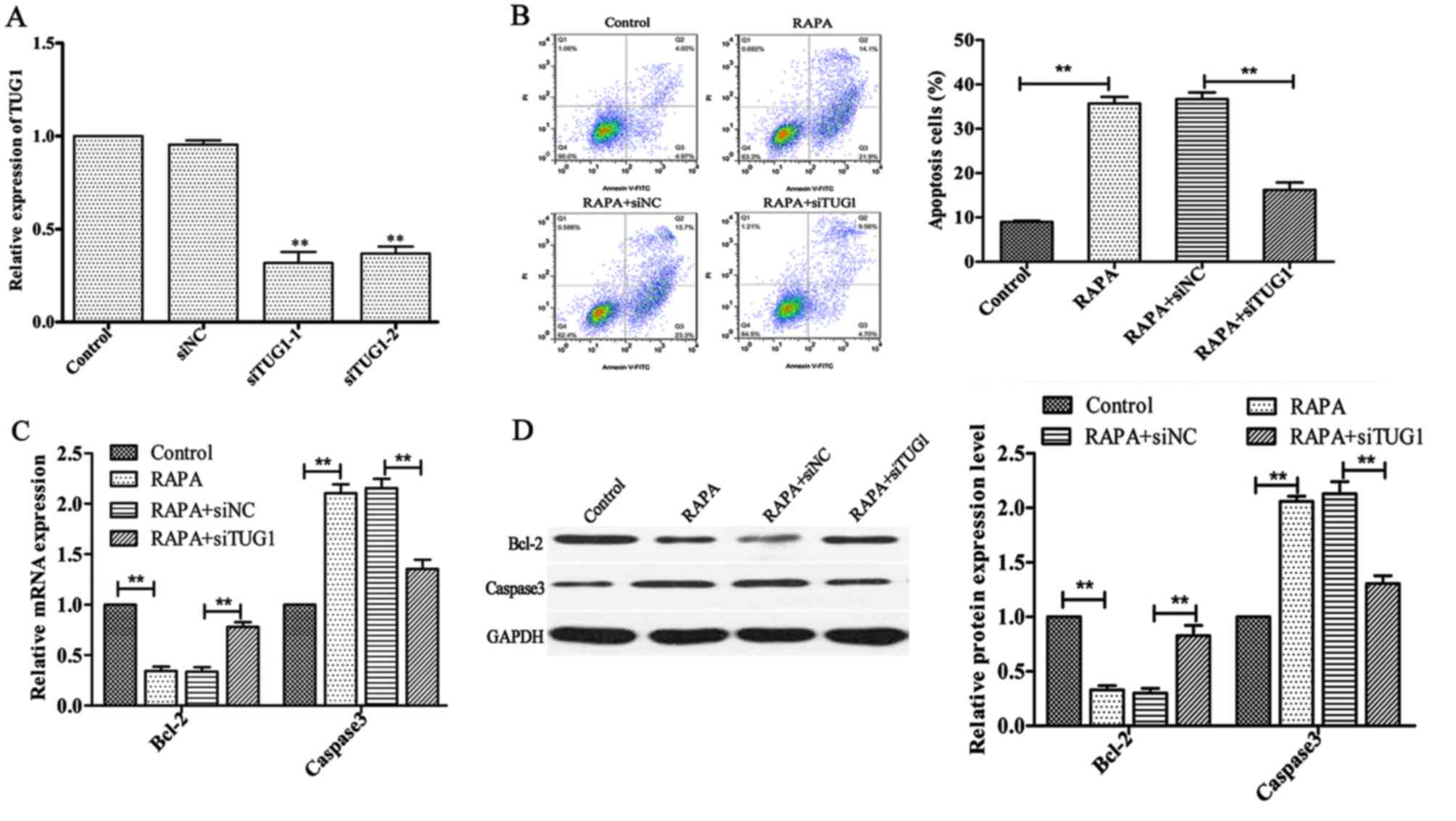
Silencing of lncRNATUG1 attenuates
rapamycin-induced apoptosis in HUVECs
To investigate whether silencing of lncRNATUG1 could
affect rapamycin-induced apoptosis in HUVECs, transfected HUVECs
were treated with 100 ng/ml rapamycin for 24 h. The results of flow
cytometry analysis demonstrated that downregulation of lncRNATUG1
expression significantly suppressed rapamycin-induced apoptosis in
HUVECs compared with the negative control (P<0.01; Fig. 4B). In addition, the expression of
Bcl-2 was revealed to be significantly upregulated, whereas the
expression of Caspase3 was revealed to be downregulated (P<0.01;
Fig. 4C and D;) following
downregulation of lncRNATUG1 compared with the negative control.
These results suggested that downregulation of lncRNATUG1 inhibits
rapamycin-induced cell apoptosis.
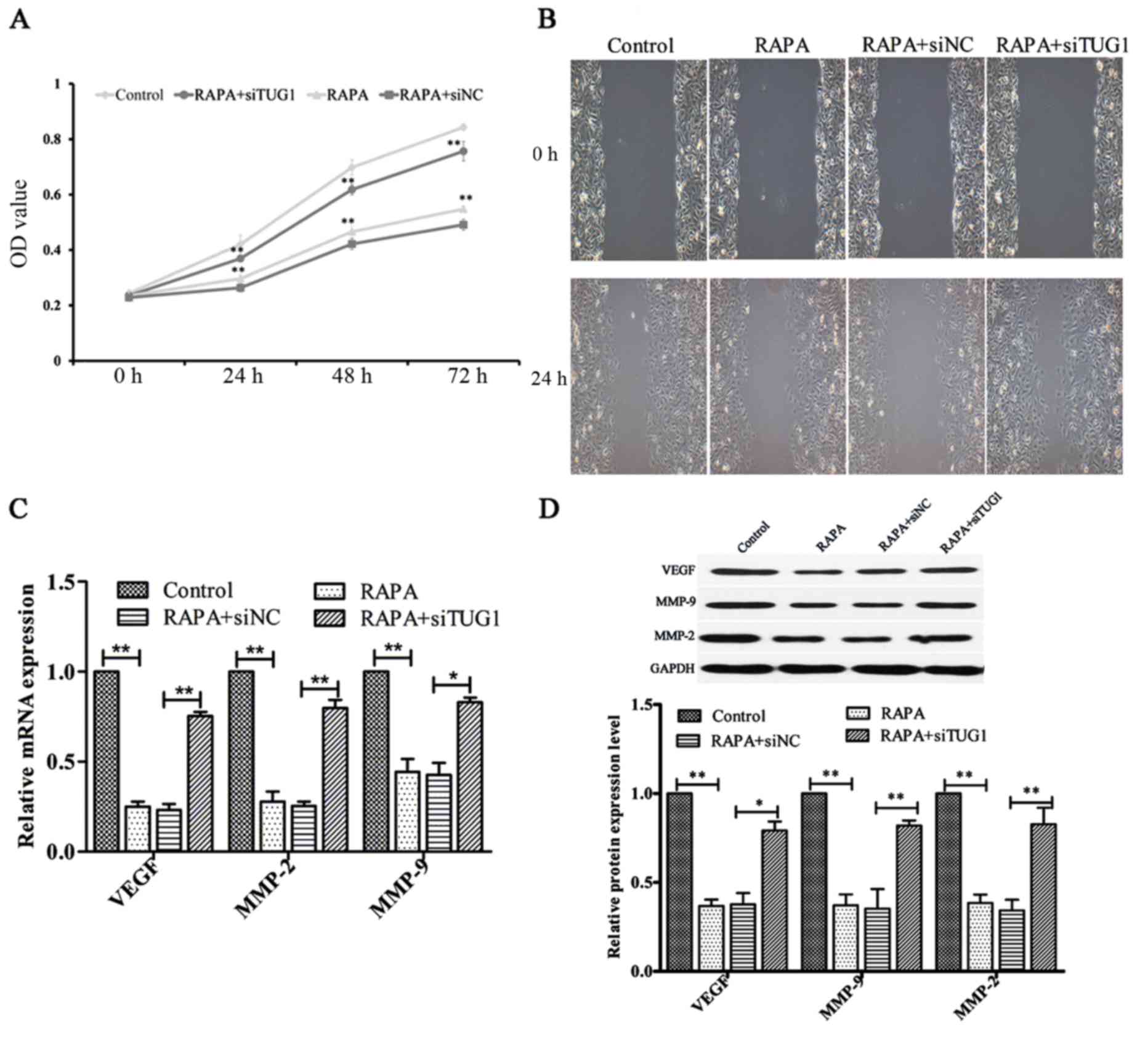
Silencing of lncRNATUG1 attenuates
rapamycin-induced inhibition of HUVEC proliferation and
migration
To investigate the effect of silencing lncRNATUG1
expression on the rapamycin-induced inhibition of HUVEC
proliferative ability, a CCK-8 assay was performed. Following
transfection, cells were treated with 100 ng/ml rapamycin for 24,
48 and 72 h. The results indicated that suppression of lncRNATUG1
expression promoted the proliferation of HUVECs compared with the
negative control (P<0.01; Fig.
5A). Additionally, subsequent wound healing analysis indicated
that downregulation of lncRNATUG1 expression increased the
migration ability of HUVECs compared with the negative control
(Fig. 5B). Furthermore, the results
of RT-qPCR and western blot analyses indicated that silencing of
lncRNATUG1 significantly upregulated the expression levels of MMP-2
(P<0.01), MMP-9 (P<0.05 for mRNA expression, P<0.01 for
protein expression) and VEGF (P<0.01 for mRNA expression,
P<0.05 for protein expression; Fig.
5C and D) compared with the negative control. These results
suggested that lncRNATUG1 may regulate rapamycin-induced inhibition
of HUVEC proliferation and migration.
Discussion
In the current study, it was demonstrated that
rapamycin suppresses the proliferation and migration of HUVECs, and
enhances the apoptosis of HUVECs, via the upregulation of
lncRNATUG1. In addition, the expression levels of VEGF, MMP-2 and
MMP-9 were indicated to be downregulated following treatment with
rapamycin. The results of the present study identified that
silencing lncRNATUG1 expression decreased the apoptosis of HUVECs
and promoted the proliferation and migration of HUVECs treated with
rapamycin; whereas the expression levels of VEGF, MMP-2 and MMP-9
were upregulated following the silencing of lncRNATUG1 expression.
Furthermore, the results of the present study revealed that the
effect of rapamycin on the proliferation, migration and apoptosis
of HUVECs may be associated with the upregulation of lncRNATUG1
expression. Therefore, lncRNATUG1 may represent a novel target for
the treatment of vascular disease.
It has been well established that the proliferation
and migration of vascular endothelial cells serves a key function
in angiogenesis (29,30). Previous studies have identified that
lncRNAs are highly expressed in endothelial cells, and are involved
in the development of cardiovascular diseases via regulation of the
proliferation and migration of endothelial cells. For example,
lincRNAp21 regulates the formation of neo-intima, as well as the
proliferation and apoptosis of VSMCs, by enhancing the
transcriptional activity of P53 and inhibiting the formation of
atherosclerosis. Therefore, lincRNA-21 may represent a therapeutic
target for atherosclerosis and other cardiovascular diseases
(31). Yan et al (32) identified that silencing lncRNA
myocardial infarction-associated transcript significantly inhibited
the proliferation and migration of vascular endothelial cells and
suppressed angiogenesis. Zhang et al (33) reported that the proliferation of
VMSCs was significantly inhibited following treatment with
baicalein and that the expression of lncRNAAK021954 was also
significantly increased. This suggests that baicalein may inhibit
the proliferation of VSMCs via regulation of lncRNAAK021954
expression.
To date, few studies have investigated lncRNATUG1
with regards to vascular disease, having instead predominantly
focused on the study of lncRNATUG1 in the context of tumorigenesis.
Previous studies have indicated that silencing of lncRNATUG1
affects the proliferation and migration of tumor cells, which
subsequently inhibits the formation of tumor blood vessels
(34,35). Zhao et al (36) indicated that silencing TUG1 with
siRNA inhibited proliferation and invasion, and promoted apoptosis,
of glioma cells. Zhang et al (37) revealed that downregulation of TUG1
inhibited proliferation, migration and invasion, and promoted
apoptosis, of renal cell carcinoma. Han et al (38) suggested that TUG1 was upregulated in
bladder cancer, and silencing TUG1 via siRNA inhibited the
proliferation and promoted apoptosis of bladder cancer cell lines.
In addition, previous studies have identified that TUG1 is highly
expressed in vascular endothelial cells (8,39,40).
Therefore, TUG1 may be involved in the regulation of vascular
endothelial cell function. Young et al (15) demonstrated that lncRNATUG1 serves a
key function in the development of the retina, and that silencing
of lncRNATUG1 expression promotes retinal cell apoptosis. These
studies suggest that TUG1 may serve a function in endothelial cell
proliferation and migration, as well as in apoptosis. In the
current study, it was revealed that, following treatment with
rapamycin, silencing of lncRNATUG1 significantly promoted the
proliferation and migration of HUVECs, as well as suppressing the
apoptosis of HUVECs. Furthermore, the expression levels of VEGF,
MMP-2 and MMP-9 were significantly upregulated following the
silencing of lncRNATUG1. The results demonstrated that lncRNATUG1
may serve a key function in endothelial cell function.
It has been widely established that rapamycin
inhibits endothelial cell proliferation and migration, and induces
apoptosis (41,42). In the current study, the results
demonstrated that rapamycin inhibits vascular endothelial cell
proliferation and migration, and reduces the expression of VEGF in
vascular endothelial cells. Rapamycin has previously been revealed
to inhibit neovascularization and the expression levels of VEGF
induced by hypoxia (43). Moss et
al (24) demonstrated that
rapamycin regulates endothelial cell migration by regulating the
cyclin-dependent kinase inhibitor p27Kip1. In addition, recent
studies have identified that lncRNAs affect the proliferation and
migration of tumor cells via regulation of the AKT/mTOR signaling
pathway (44–46). The results of the present study
demonstrated that rapamycin significantly inhibited the
proliferation and migration of HUVECs, and enhanced the apoptosis
and expression of lncRNATUG1 in HUVECs. However, silencing of TUG1
expression significantly inhibited rapamycin-induced apoptosis and
promoted endothelial cell proliferation and migration. In addition,
the expression levels of VEGF, MMP-2 and MMP-9 were upregulated in
HUVECs treated with rapamycin following silencing of TUG1
expression. These results indicate that lncRNATUG1 affects
endothelial cell function and may be involved in the suppression of
the proliferation and migration of HUVECs, as well as the promotion
of the apoptosis of HUVECs, following treatment with rapamycin.
In conclusion, the results of the present study
revealed that rapamycin suppresses the proliferation and migration
of HUVECs, and enhances the apoptosis of HUVECs, via the lncRNATUG1
pathway. This suggests that lncRNATUG1 serves a key function in
rapamycin-induced inhibition of endothelial cell proliferation and
migration, and may represent a novel therapeutic target for the
treatment of vascular stenosis. However, the exact regulatory
mechanism of lncRNATUG1 requires further study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Health
Science and Technology Project of Yunnan Province (grant nos.
2016NS188 and 2018NS0008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XG and YY conceived and designed the current study;
XG, TZ, XZ and GL performed the experiments and analyzed the data;
XG wrote the manuscript; and LD, ZM and JW assisted with performing
the experiments and analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qureshi 1A, Mattick JS and Mehler MF: Long
non-coding RNAs in nervous system function and disease. Brain Res.
1338:20–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dinger ME, Amaral PP, Mercer TR, Pang KC,
Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C,
et al: Long noncoding RNAs in mouse embryonic stem cell
pluripotency and differentiation. Genome Res. 18:1433–1445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kanduri C: Long noncoding RNA and
epigenomics. Adv Exp Med Biol. 722:174–195. 2015. View Article : Google Scholar
|
|
6
|
Yin DD, Zhang E B, You L H, Wang N, Wang
LT, Jin FY, Zhu YN, Cao LH, Yuan QX, De W and Tang W:
Down-regulation of lncRNA TUG1 affects apoptosis and insulin
secretion in mouse pancreatic β cells. Cell Physiol Biochem.
35:1892–1904. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma
S and Wang YJ: Role of LncRNA TUG1 in intervertebral disc
degeneration and nucleus pulposus cells via regulating
Wnt/β-catenin signaling pathway. BiochemBiophys Res Commun.
491:668–674. 2017. View Article : Google Scholar
|
|
8
|
Michalik KM, You X, Manavski Y,
Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W,
Uchida S, et al: Long noncoding RNA MALAT1 regulates endothelial
cell function and vessel growth. Circ Res. 114:1389–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang M, Gu H, Xu W and Zhou X:
Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis
and improves left ventricular function in diabetic rats. Int J
Cardiol. 203:214–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5:e15062014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tao H, Cao W, Yang JJ, Shi KH, Zhou X, Liu
LP and Li J: Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in
cardiac fibroblast proliferation and fibrosis. Cardiovasc Pathol.
25:381–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pan JX: LncRNA H19 promotes
atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur
Rev Med Pharmacol Sci. 21:322–328. 2017.PubMed/NCBI
|
|
13
|
Ballantyne MD, Pinel K, Dakin R, Vesey AT,
Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et
al: Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates
Cell Proliferation. Circulation. 133:2050–2065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu GZ, Tian W, Fu HT, Li CP and Liu B:
Long noncoding RNA-MEG3 is involved in diabetes mellitus-related
microvascular dysfunction. Biochem Biophys Res Commun. 471:135–141.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan S, Yang Z, Ke Z, Huang K, Liu N, Fang
X and Wang K: Down-regulation of the long non-coding RNA TUG1 is
associated with cell proliferation, migration, and invasion in
breast cancer. Biomed Pharmacother. 95:1636–1643. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yun-Bo F, Xiao-Po L, Xiao-Li L, Guo-Long
C, Pei Z and Fa-Ming T: LncRNA TUG1 is up-regulated and promotes
cell proliferation in osteosarcoma. Open Med (Wars). 11:163–167.
2016.PubMed/NCBI
|
|
18
|
Rosner D, Mccarthy N and Bennett M:
Rapamycin inhibits human in stent restenosis vascular smooth muscle
cells independently of pRB phosphorylation and p53. Cardiovasc Res.
66:601–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawatsu S, Oda KY, Tabata Y and Tabayashi
K: External application of rapamycin-eluting film at anastomotic
sites inhibits neointimal hyperplasia in a canine model. Ann Thorac
Surg. 84:560–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Heijden DJ, Westendorp IC,
Riezebos RK, Kiemeneij F, Slagboom T, van der Wieken LR and Laarman
GJ: Lack of efficacy of clopidogrel pre-treatment in the prevention
of myocardial damage after elective stent implantation. J Am Coll
Cardiol. 44:20–24. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matetzky S, Shenkman B, Guetta V, Shechter
M, Bienart R, Goldenberg I, Novikov I, Pres H, Savion N, Varon D
and Hod H: Clopidogrel resistance is associated with increased risk
of recurrent atherothrombotic events in patients with acute
myocardial infarction. Circulation. 109:3171–3175. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gurbel PA, Bliden KP, Samara W, Yoho JA,
Hayes K, Fissha MZ and Tantry US: Clopidogrel effect on platelet
reactivity in patients with stent thrombosis: Results of the CREST
study. J Am Coll Cardiol. 46:1827–1832. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geisler T, Langer H, Wydymus M, Göhring K,
Zürn C, Bigalke B, Stellos K, May AE and Gawaz M: Low response to
clopidogrel is associated with cardiovascular outcome after
coronary stent implantation. Eur Heart J. 27:2420–2425. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moss SC, Lightell DJ Jr, Marx SO, Marks AR
and Woodset TC: Rapamycin Regulates Endothelial Cell Migration
through Regulation of the Cyclin-dependent Kinase Inhibitor
p27Kip1. J Biol Chem. 285:11991–11997. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li E, Zhao Z, Ma B and Zhang J: Long
noncoding RNA HOTAIR promotes the proliferation and metastasis of
osteosarcoma cells through the AKT/mTOR signaling pathway. Exp Ther
Med. 14:5321–5328. 2017.PubMed/NCBI
|
|
26
|
Wang Y, Wang Y, Li J, Zhang Y, Yin H and
Han B: CRNDE, a long-noncoding RNA, promotes glioma cell growth and
invasion through mTOR signaling. Cancer Lett. 367:122–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsumoto A, Pasut A, Matsumoto M,
Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI,
Clohessy JG and Pandolfi PP: mTORC1 and muscle regeneration are
regulated by the LINC00961-encoded SPAR polypeptide. Nature.
541:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muñoz-Chápuli R, Quesada AR and Medina
Angel M: Angiogenesis and signal transduction in endothelial cells.
Cell Mol Life Sci. 61:2224–2243. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lamalice L, Le Boeuf F and Huot J:
Endothelial cell migration during angiogenesis. Circ Res.
100:782–794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen
C, Cai Y, Huang H, Yang Y, Liu Y, et al: LincRNA-p21 Regulates
neointima formation, vascular smooth muscle cell proliferation,
apoptosis and atherosclerosis by enhancing p53 activity.
Circulation. 130:1452–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan B, Yao J, Liu J Y, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Ma G, Li C, Cao Z, Qie F and Xu
X: Baicaleininhibits VSMCs proliferation via regulating
LncRNAAK021954 gene expression. Int J Clin Exp Med. 8:22129–22138.
2015.PubMed/NCBI
|
|
34
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Y, Sun X, Mao C, Guo G, Ye S, Xu J, Zou
R, Chen J, Wang L, Duan P and Xue X: Upregulation of long noncoding
RNA TUG1 promotes cervical cancer cell proliferation and migration.
Cancer Med. 6:471–482. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Z, Wang B, Hao J, Man W, Chang Y, Ma
S, Hu Y, Liu F and Yang J: Downregulation of the long non-coding
RNA taurine-upregulated gene 1 inhibits glioma cell proliferation
and invasion and promotes apoptosis. Oncol Lett. 15:4026–4032.
2018.PubMed/NCBI
|
|
37
|
Zhang M, Lu W, Huang YQ, Shi JZ, Wu X,
Zhang XL, Jiang RZ, Cai ZM and Wu S: Downregulation of the long
noncoding RNA TUG1 inhibits the proliferation, migration, invasion
and promotes apoptosis of renal cell carcinoma. J Mol Histol.
47:421–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han YH, Liu YC, Gui YT and Cai ZM: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y,
Li Z, Shang X and Liu Y: The long noncoding RNA TUG1 regulates
blood-tumor barrier permeability by targeting miR-144. Oncotarget.
6:19759–19779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen C, Cheng GQ, Yang XN, Li CS, Shi R
and Zhao NN: Tanshinol suppresses endothelial cells apoptosis in
mice with atherosclerosis via lncRNA TUG1 up-regulating the
expression of miR-26a. Am J Transl Res. 8:2981–2991.
2016.PubMed/NCBI
|
|
41
|
Liu HT, Li F, Wang WY, Li XJ, Liu YM, Wang
RA, Guo WY and Wang HC: Rapamycin inhibits re-endothelialization
after percutaneous coronary intervention by impeding the
proliferation and migration of endothelial cells and inducing
apoptosis of endothelial progenitor cells. Tex Heart Inst J.
37:194–201. 2010.PubMed/NCBI
|
|
42
|
Wang Y, Chen J, Tang W, Zhang Y and Li X:
Rapamycin inhibits the proliferation of endothelial cells in
hemangioma by blocking the mTOR-FABP4 pathway. Biomed Pharmacother.
85:272–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Medici D and Olsen BR: Rapamycin inhibits
proliferation of hemangioma endothelial cells by reducing
HIF-1-dependent expression of VEGF. PloS One. 7:e429132012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xue D, Zhou C, Lu H, Xu R, Xu X and He X:
LncRNA GAS5 inhibits proliferation and progression of prostate
cancer by targeting miR-103 through AKT/mTOR signaling pathway.
Tumor Biol. 37:1–11. 2016. View Article : Google Scholar
|
|
45
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long Noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 up-regulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shui X, Zhou C, Lin W, Yu Y, Feng Y and
Kong J: Long non-coding RNA BCAR4 promotes chondrosarcoma cell
proliferation and migration through activation of mTOR signaling
pathway. Exp Biol Med (Maywood). 242:1044–1050. 2017. View Article : Google Scholar : PubMed/NCBI
|