Introduction
Hepatitis B virus (HBV) is a pandemic disease, with
an estimated 240 million individuals harboring Hepatitis B surface
antigen (HBsAg). Patients with chronic hepatitis B (CHB) are at an
increased risk of developing cirrhosis and hepatocellular
carcinoma. At present, treatments for CHB include pegylated
interferon and nucleos(t)ide analogues (NAs) (1). Among these, lamivudine (LAM), adefovir
dipivoxil (ADV), entecavir (ETV), telbivudine (LdT), tenofovir
disoproxil fumarate (TDF) and tenofovir alafenamide have been
approved for CHB treatment (1).
Hepatitis B e antigen (HBeAg) seroconversion is considered to be
the most important marker for assessing the reliability and
efficacy of antiviral therapy in patients with HBeAg-positive CHB
(2).
MicroRNAs (miRNAs or miRs) are small (19–24
nucleotides) non-coding RNA molecules that regulate a variety of
cellular processes (3). A number of
miRNAs have been reported to participate in the regulation of HBV
infection and related diseases (4–7). A
previous study demonstrated that serum miR-125b was correlated with
HBV replication and liver necroinflammation (8), while others have demonstrated that
miR-125b-5p expression is associated with the etiology of chronic
HBV infection and regulates HBsAg expression (9–10). it
has also been observed that miR-125b inhibits the formation of HBV
DNA intermediates and the secretion of HBsAg and HBeAg by targeting
sodium channel epithelial 1α subunit (11). The various functions of miR-125b in
HBV-associated liver diseases suggest that miR-125b level may have
clinical value.
Previous studies have identified multiple predictors
of NA treatment outcome, including pre-treatment alanine
aminotransferase (ALT) level, HBV DNA level during treatment and
HBsAg, HBeAg and anti- Hepatitis B core antigen (HBcAg) antibody
levels (12–19); however, the predictive value of serum
miR-125b level for NA treatment response is unknown. The aim of the
present study was to assess whether miR-125b expression, alone or
in combination with other parameters, is an effective predictor of
complete remission (CR) following 44 weeks of Nas therapy (CR is
defined as HBV DNA <500 IU/ml and HBeAg seroconversion) in
patients with CHB.
Patients and methods
Patients
A total of 66 HBeAg-positive CHB patients [age
range, 17–64 years; males, 84.8% (56/66)] were retrospectively
analyzed. All patients had received optimized LdT therapy (with
additional ADV if HBV DNA ≥500 IU/ml at week 24; n=39) or TDF
monotherapy (n=27) for at least 144 weeks at the Department of
Infectious Diseases, Huashan Hospital (Fudan University, Shanghai,
China) between January 2013 and December 2016. A total of 34
healthy individuals (age range, 18–60 years) matched for age and
with 85.3% (29/34) males were enrolled as healthy controls (HCs) at
Huashan Hospital. Inclusion criteria were as follows: Baseline
serum samples available, aged 16–65 years, HBsAg positive for a ≥6
months, HBeAg positive, anti-HB e antibody (anti-HBe) negative;
serum HBV DNA ≥20,000 IU/ml; ALT ≥2× upper limit of normal (ULN)
and no history of antiviral therapy with NA or interferon within
the previous 6 months. Exclusion criteria were as follows:
Simultaneously positive for HBeAg and anti-Hbe, co-infection with
hepatitis C virus, hepatitis D virus, or human immunodeficiency
virus, hepatic decomposition and history of other acquired or
inherited causes of liver disease. All patients provided written,
informed consent prior to participation in the study. The study
protocol was performed in accordance with the Declaration of
Helsinki and was approved by the Institutional Ethics Committee of
Huashan Hospital.
Evaluation of miR-125b levels and
other serological parameters
Quantitative miR-125b evaluation was performed for
all patients at baseline. Serum miRNA isolation and quantification
was performed using miRcute miRNA extraction and first-strand cDNA
synthesis and qPCR detection kits (cat. nos. DP503 and KR201;
Tiangen Biotech Co., Ltd., Beijing, China) according to the
manufacturer's protocol. All reactions were performed in
triplicate. The miRNA levels were normalized to an internal control
(5S rRNA) according to the manufacturer's protocol. Primers for 5S
(cat. no. 201-0001) and miR-125b (cat. no. 201-00047) were
purchased from Tiangen Biotech Co., Ltd and the relative expression
of target miRNAs was determined using the 2−∆∆Cq method
(20).
HBV DNA levels were determined using the Da-an
real-time PCR HBV DNA assay (cat. no. DA-L051; Daan Gene Co, Ltd of
Sun Yat-sen University, Guangdong, China) following the
manuscript's instrument. According to the instructions, HBV DNA was
extracted from 100 µl serum. The TaqMan probe (provided as part of
the kit) was used in qPCR amplification, performed using the
LightCycler 480 system (Roche Diagnostics, Basel, Switzerland), by
incubating the reaction mixture at 93°C for 2 min, followed by 40
cycles of 93°C for 5 sec and 57°C for 45 sec. The dynamic range for
this kit to detect HBVDNA ranged from
5×102−1×108 IU/ml.
The quantification of HBsAg was performed by ADICON
Clinical Laboratories (Shanghai, China) using the Abbott ARCHITECT
I2000 platform (Abbott Pharmaceutical Co. Ltd., Lake Bluff, IL,
USA). Serological HBV markers were measured by Chemiluminescent
Microparticle ImmunoAssays for HBsAg, anti-HBs, HBeAg and anti-HBe
(cat. nos. 6C36, 7C18, 6C32 and 6C34; Abbott Pharmaceutical Co.
Ltd.). Serum HBsAg and anti-HBs were determined quantitatively,
while serum HBeAg and anti-HBe were determined qualitatively.
Positive cut-offs values for HBsAg and anti-HBs were ≥0.05 and ≥10
IU/l, respectively. The upper detection limits for HBsAg and
anti-HBs were 250 and 1,000 IU/l, respectively. HBeAg and anti-HBe
were interpreted using a ratio of the sample relative light unit
(RLU) rate/the cut-off RLU.
Statistical analysis
Data were analyzed using SPSS v.19.0 for Windows
(IBM Corp., Armonk, NY, USA). Categorical and continuous variables
are presented as a proportion (%) and median (range), respectively.
Pearson's χ2 analysis or Fisher's exact test were used
to compare categorical variables, while the Student's t-test or
Wilcoxon non-parametric test was used for normally distributed
data. Cumulative CR rates were analyzed with the Kaplan-Meier
method and significant differences were determined using the
log-rank test. Variables with P values <0.10 were subjected to
multivariate logistic regression analysis to identify independent
variables for predicting CR. The optimal cut-off value of each
variable was determined by the Youden index using MedCalc v.4.20
(MedCalc Software, Mariakerke, Belgium). Serum HBsAg and HBV DNA
levels are expressed as log values. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics and treatment
outcomes
It was observed that the serum miRNA was positively
correlated with serum ALT (r=0.300; P=0.014) and HBV DNA (r=0.353;
P=0.004) prior to treatment (Fig.
1). Furthermore, it was suggested that in the CHB group, serum
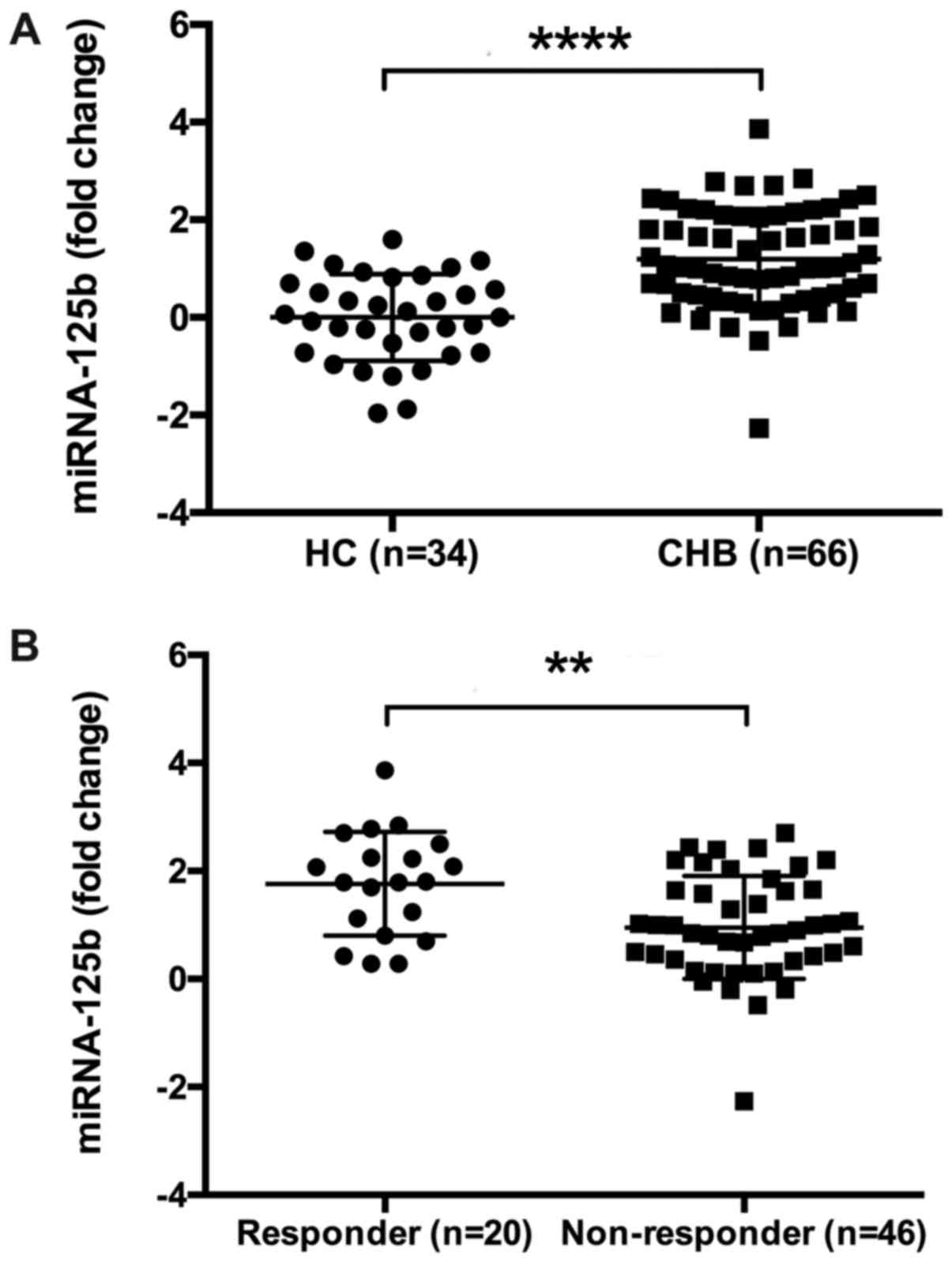
miRNA-125b levels were significantly higher compared with HCs
(P=0.0037; Fig. 2A). A total of 66
patients, who received LdT optimized therapy (n=39; 12 patients
supplemented with ADV during the treatment) or TDF monotherapy
(n=27) were analyzed. The baseline characteristics of patients who
achieved or did not achieve CR following 144 weeks of NA treatment
are presented in Table I. Baseline
fold change in serum miR-125b (1.80 vs. 0.87; P=0.002; Fig. 2B) were significantly higher in the CR
group compared with the non-CR group, while HBsAg (3.88 vs. 4.46
log10 IU/ml; P=0.009) levels and percentages of ALT <5× ULN (11%
vs. 37%; P=0.033) at baseline were significantly lower in CR group
compared with the non-CR group. No significant differences were
observed with respect to age, sex, baseline serum HBV DNA level,
HBeAg level or HBV genotype (Table
I). The cumulative CR rates of patients who received LdT
optimized therapy were 23.1 and 35.9% at weeks 96 and 144,
respectively (Fig. 3). The 144-week
cumulative CR rate of patients who received TDF monotherapy was
22.2%.
 | Table I.Baseline characteristics of
responders and non-responders. |
Table I.
Baseline characteristics of
responders and non-responders.
| Characteristic | All patients
(n=66) | Responders
(n=20) | Non-responders
(n=46) | P-value |
|---|
| Age (years) | 30 (17–64) | 28 (20–60) | 31 (17–64) | 0.785 |
| Male sex, n
(%) | 56 (84.8) | 17 (85.0) | 39 (84.8) | 1.000 |
| Nucleos(t)ide
analogue |
|
|
|
|
| LdT
optimized therapy, n (%) | 39 (59.1) | 14 (70.0) | 25 (54.3) |
|
| TDF
monotherapy, n (%) | 27 (40.9) | 6 (30.0) | 21 (45.7) | 0.235 |
| Serum miR-125b
(fold change) | 1.02
(−2.27–3.86) | 1.80
(0.29–3.86) | 0.87
(−2.27–2.70) | 0.002 |
| ALT (ULN) | 2.96
(0.48–16.74) | 3.96
(0.50–11.14) |
2.93(0.48–16.74) | 0.410 |
| ≤5, n
(%) | 48 (72.7) | 11 (55.0) | 37 (80.4 |
|
| >5,
n (%) | 18 (27.3) | 9 (45.0) | 9 (19.6) | 0.033 |
| Log10
HBV DNA (IU/ml) | 7.60
(4.00–9.76) | 7.64
(4.00–9.38) | 7.60
(4.00–9.76) | 0.751 |
| <8,
n (%) | 41 (62.1) | 13 (65.0) | 28 (60.9) |
|
| ≥8, n
(%) | 25 (37.9) | 7 (35.0) | 18 (39.1) | 1.000 |
| Log10
HBeAg (s/co) | 2.92
(0.48–3.64) | 2.54
(0.90–3.64) | 3.04
(0.48–3.23) | 0.075 |
| Log10
HBsAg (IU/ml) | 4.28
(2.00–5.28) | 3.88
(2.00–4.88) | 4.46
(2.72–5.28) | 0.009 |
|
<4.4, n (%) | 38 (57.6) | 16 (80.0) | 22 (47.8) |
|
| ≥4.4, n
(%) | 28 (42.4) | 4 (20.0) | 24 (52.2) | 0.015 |
| HBV genotype
(%) |
|
|
|
|
| B | 38 (57.6) | 12 (60) | 26 (56.5) |
|
| C | 28 (42.4) | 8 (40) | 20 (43.4) | 0.945 |
Baseline and on-treatment parameters
associated with 144-week CR
To identify factors associated with CR, baseline
parameters including age, sex, treatment strategy, ALT >5× ULN,
HBV DNA level, HBsAg level, HBeAg level and serum miR-125b level
were included in the logistic regression analysis, as were
on-treatment parameters, including undetectable HBV DNA (HBV DNA
<500 IU/ml) at week 24. Uni- and multivariate analyses
demonstrated that changes in serum miR-125b (OR=4.377; P=0.006),
HBsAg (OR=0.120; P=0.010), ALT >5× ULN (OR=11.726; P=0.018) and
undetectable HBV DNA (OR=7.828; P=0.021) between the baseline and
week 24 were independent predictors of 144-week CR (Table II).
 | Table II.Logistic regression analysis of
parameters to predict complete response at week 144. |
Table II.
Logistic regression analysis of
parameters to predict complete response at week 144.
|
| Complete
response |
|---|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factors | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Age (years) | 0.994
(0.950–1.040) | 0.781 |
|
|
| Male sex, n
(%) | 1.017
(0.234–4.413) | 0.982 |
|
|
| Baseline |
|
|
|
|
| TDF
monotherapy | 0.510
(0.167–1.561) | 0.238 | 0.516
(0.105–2.542) | 0.416 |
| ALT
>5 ULN | 3.364
(1.072–10.550) | 0.038 | 11.726
(1.512–90.920) | 0.018 |
|
Log10 HBV DNA
(IU/ml) | 0.886
(0.632–1.242) | 0.482 |
|
|
|
Log10 HBeAg
(s/co) | 0.601
(0.339–1.063) | 0.080 | 1.379
(0.472–4.034) | 0.557 |
|
Log10 HBsAg
(IU/ml) | 0.391
(0.184–0.832) | 0.015 | 0.120
(0.024–0.597) | 0.010 |
| Serum
miR-125b (fold change) | 2.561
(1.332–4.927) | 0.005 | 4.377
(1.513–12.661) | 0.006 |
| Week 24 |
|
|
|
|
| HBV DNA
<500 (IU/ml) | 7.424
(2.3–23.969) | 0.001 | 7.828
(1.371–44.711) | 0.021 |
Value of baseline HBsAg and miR-125b
fold change for predicting 144-week CR
The predictive value of baseline HBsAg and miR-125b
fold change for 144-week CR was evaluated by calculating the area
under the receiver operating characteristic curve (AUROC). The
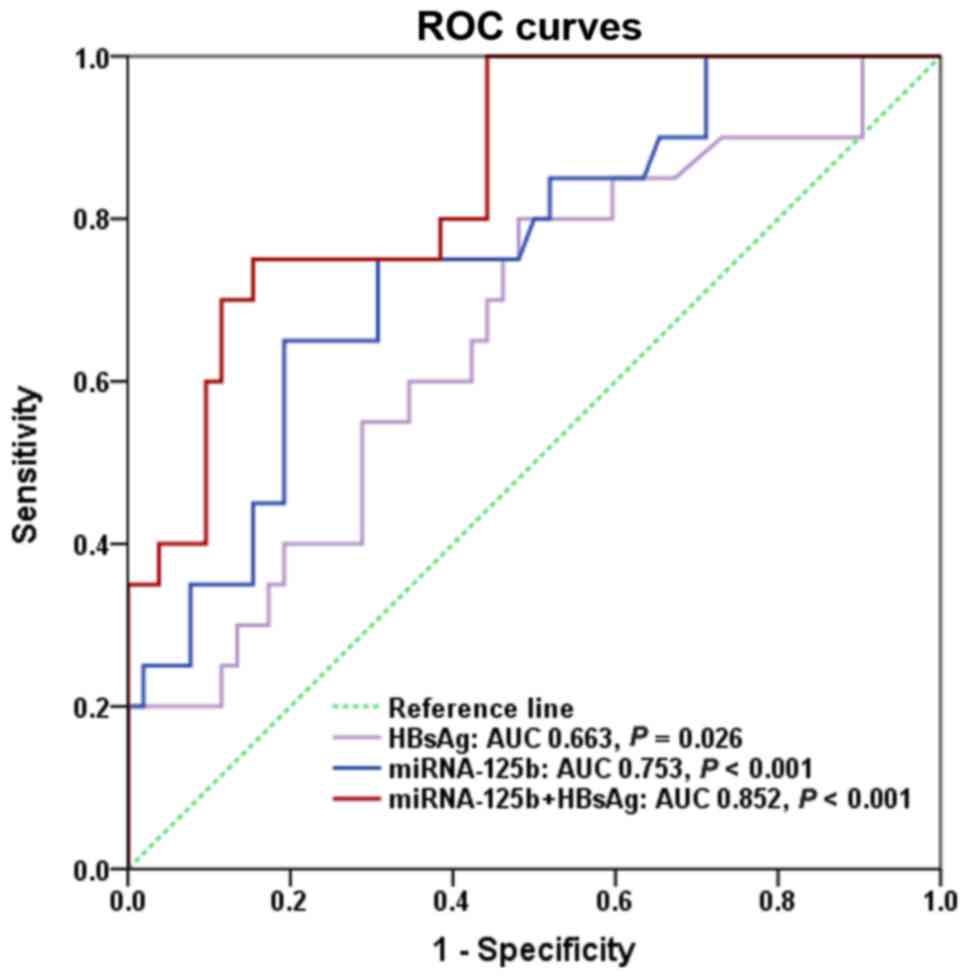
AUROC of miR-125b at the baseline was 0.753 (P<0.001). Baseline
HBsAg levels were revealed to predict CR at week 144 with an AUROC
of 0.663 (P=0.026). The combination of miR-125b fold change and
baseline HBsAg level at the baseline had a higher predictive value
than either parameter alone, with an AUROC of 0.852 (P<0.001;
Fig. 4). Based on ROC analysis,
several cut-off values of fold change in serum miR-125b and HBsAg
level at baseline were selected for further evaluation. A cut-off
value of 1.7 was considered optimal for predicting 144-week CR
based on fold change in baseline miR-125b, with a sensitivity of
65%, specificity of 78.3%, positive predictive value (PPV) of 56.5%
and negative predictive value (NPV) of 83.7%. The optimal cut-off
value for baseline HBsAg (log10 IU/ml) was 4.4, with a
sensitivity of 80%, specificity of 52.2%, PPV of 42.1% and NPV of
85.7% (Table III).
 | Table III.Predictive value of miRNA-125b fold
change and HBsAg levels for complete response at 144 weeks. |
Table III.
Predictive value of miRNA-125b fold
change and HBsAg levels for complete response at 144 weeks.
| Cut-off values | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) |
|---|
| miRNA-125b (fold
change) |
|
1.0 | 75.0 | 56.5 | 42.9 | 83.9 |
|
1.3 | 65.0 | 67.4 | 46.4 | 81.6 |
|
1.7a | 65.0 | 78.3 | 56.5 | 83.7 |
|
2.1 | 35.0 | 82.6 | 46.7 | 74.5 |
|
2.4 | 25.0 | 93.5 | 62.5 | 74.1 |
| Log10
HBsAg |
|
|
|
|
|
4.1 | 60.0 | 63.0 | 41.4 | 78.4 |
|
4.3 | 70.0 | 56.5 | 41.2 | 81.2 |
|
4.4a | 80.0 | 52.2 | 42.1 | 85.7 |
|
4.6 | 85.0 | 41.3 | 38.6 | 86.4 |
|
4.7 | 90.0 | 30.4 | 36.0 | 87.5 |
| Combination | 45.0 | 92.3 | 69.2 | 79.2 |
Predictive algorithms for predicting
144-week CR
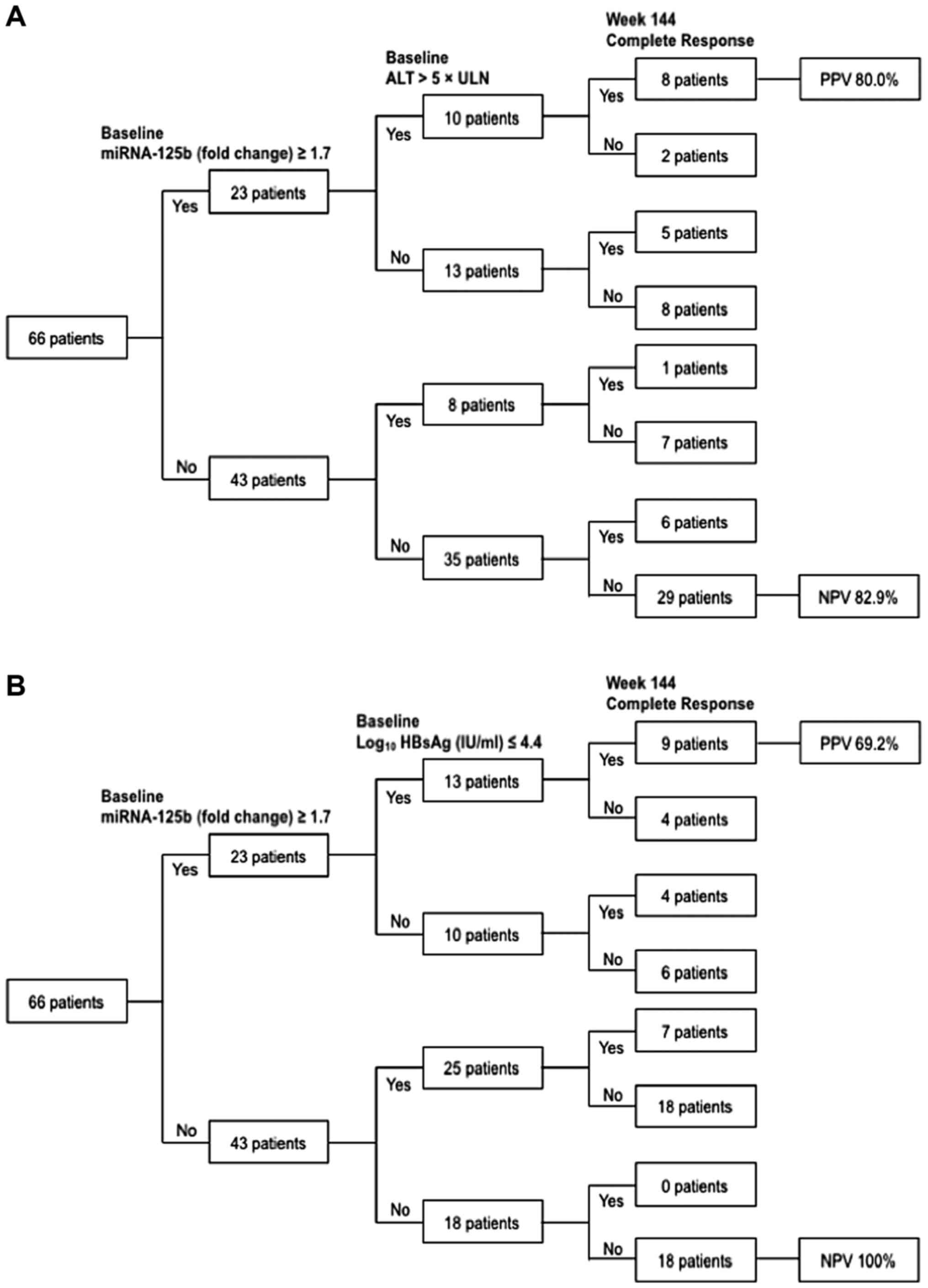
Predictive algorithms for CR at week 144 were
developed based on baseline serum miR-125b combined with ALT or
HBsAg levels (Fig. 5). A total of 10
patients achieved miRNA-125b ≥1.7 and ALT >5× ULN at the
baseline. A total of 80% (8/10) patients achieved CR by the end of
week 144 (PPV=80%; Fig. 5A).
However, 18 patients did not achieve miR-125b ≥1.7 and HBsAg ≤4.4
(log10 IU/ml) at the baseline. None of these patients achieved CR
by the end of week 144 (NPV=100%; Fig.
5B).
Discussion
HBeAg seroconversion is an important landmark in the
treatment of patients with HBeAg-positive CHB (2). Given the indeterminate duration of NA
treatment and the drawbacks of long-term therapy (1), predicting HBeAg seroconversion has
clinical benefits.
A number of virological parameters have been
investigated for their utility as predictors of NA treatment
efficacy in patients with CHB. Serum HBV DNA at week 24 has been
reported to be an essential marker for monitoring HBeAg status in
HBeAg-positive patients receiving long-term NA treatment (21). The combination of HBeAg levels at
baseline and its decline from baseline after 24 weeks may be a
useful predictor of the efficacy of NA therapy (15). Baseline anti-HBcAg titer has also
been used to predict the therapeutic efficacy of NAs in
HBeAg-positive CHB patients (16),
while patients with low HBsAg levels demonstrated satisfactory
responses to NA treatment (17–18). In
addition to these virological parameters, HBeAg seroconversion
following NA treatment can be predicted by high baseline ALT
(12) and serum interleukin-21
levels at week 12 (22). Quantifying
interferon-γ-inducible protein-10 during entecavir (ETV) treatment
may predict long-term HBeAg seroconversion in patients with CHB
(23).
Changes in miRNA expression have been linked to
disease progression in patients with HBV (24). Serum miR-125b has previously been
reported to be associated with HBV replication, liver
necroinflammation (8) and the
etiology of CHB infection (via regulating BsAg expression)
(10). miR-125b inhibits the
formation of HBV DNA intermediates and HBsAg and HBeAg secretion
(11). miR-125b can also regulate
several oncogenes, including Mothers against decapentaplegic
homolog 2/4), Sirtuin 7, suppressor of variegation 3–9 homolog 1,
Lin-28 homolog B and phosphatidylinositol-glycan biosynthesis class
F protein (25–29). These genes are associated with HBV
hepatocarcinogenesis, which indicates that miR-125b also serves an
important role in HBV hepatocarcinogenesis.
Multivariate regression analysis identified baseline
miR-125b level as an independent predictor of 144-week CR. It was
also demonstrated that HBsAg level, ALT >5× ULN at the baseline
and HBV DNA <500 IU/ml at week 24 were independently associated
with 144-week CR, which is consistent with previous studies
(16). The AUROC of anti-HBcAg
antibody at the baseline was reported as 0.646 for predicting HBeAg
seroconversion following NA treatment; however, the AUROC of
baseline miR-125b level for predicting CR was 0.753 and increased
to 0.852 when combined with HBsAg level.
Cut-off values for miR-125b and HBsAg were obtained
via ROC analysis. The sum of sensitivity and specificity was
highest when the cut-off values were 1.7 for miR-125b fold change
and 4.4 log10 IU/ml for HBsAg. The predictive algorithm
combining fold change in baseline miR-125b ≥1.7 and HBsAg ≤4.4
(log10 IU/ml) demonstrated good performance in excluding
144-week CR following NA treatment, with an NPV of 100%. This
indicates that patients who did not exhibit a fold change ≥1.7 in
miR-125b and ≤4.4 in HBsAg (log10 IU/ml) at the baseline
would have little chance of achieving CR with long-term NA therapy.
Another predictive algorithm, combining ≥1.7 fold change in
baseline miR-125b and ALT >5× ULN, was a good predictor of
144-week CR, with a PPV of 80%. These results suggest that a
patient achieving ≥1.7 fold change in miR-125b and ALT >5× ULN
at the baseline is likely to respond favorably to long-term NA
therapy.
The majority of patients enrolled in the present
study were <40 years of age; previous studies recommended that
young patients with CHB and fertility requirements should receive
LdT and TDF as a first-choice treatment, as these agents have ben
reported to reduce perinatal HBV transmission (30,31). Two
global phase III clinical trials of NA TDF reported that HBeAg
seroconversion rates were 21 and 40% at weeks 48 and 240,
respectively, in patients with HBeAg-positive CHB (32) LdT is an L-deoxythymidine analogue
that has greater efficacy than LAM in patients with CHB (33). Although LdT is not recommended as a
first-line NA according to current guidelines, it is widely
prescribed to patients with CHB in developing countries due to its
low cost and potency, which is comparable to that of ETV and TDF
(34,35). The rate of HBeAg seroconversion was
29.6% in a 2-year global trial of LdT (36). However, cumulative HBeAg
seroconversion rates in the present study were lower than those
previously reported, likely due to the small sample size and
predominance of HBV genotype C in China, which has lower rates of
seroconversion than genotypes A and B (37).
To the best of our knowledge, the present study is
the first to report the predictive value of baseline serum miR-125b
for patient response to long-term NA treatment. However, a number
of limitations were noted. Firstly, the sample size and age range
of enrolled patients relatively small, and so the conclusions
herein require confirmation in multi-center studies with a large
cohort. Secondly, all patients enrolled were Asian; as such, it is
unclear whether the results can be generalized to other ethnic
groups or HBV genotypes. Thirdly, the present study lacked a
validation group in which to verify the predictive model. In the
future, a prospective cohort should be enrolled to evaluate and
confirm the predictive validity of miR-125b at the baseline and
during therapy. Nonetheless, the present study suggests that
baseline miR-125b is a reliable predictor for HBeAg seroconversion
following NA treatment. These results may be used in the clinic to
optimize therapeutic regimens prior to initiating antiviral
treatment.
Acknowledgements
The authors would like to thank Dr. Xiaoqin Wang
from the Department of Hemotology Diseases at Huashan Hospital for
the assistance during the study.
Funding
The present study was supported by the Science and
Technology Commission of Shanghai Municipality (grant no.
17ZR1403600), the Three-year Action Plan on Public Health, Phase
IV, Shanghai, China (grant no. 15GWZK0801) and the National Natural
Science Foundation of China (grant nos. 81670560 and 1471933).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PZ, MD, JZ and JG were responsible for conception,
study design and data analysis. PZ, MD, JW FL, JZ and JG performed
data collection. PZ and MD wrote the manuscript. JZ and JG
critically revised the manuscript prior to submission. All authors
approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were approved by the Ethic Committee of Huashan
Hospital, Fudan University and in accordance with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards. Informed consent was obtained from all individual
participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interest.
Glossary
Abbreviations
Abbreviations:
|
ADV
|
adefovir dipivoxil
|
|
ALT
|
alanine aminotransferase
|
|
AUROC
|
area under the receiver operating
characteristic curve
|
|
CHB
|
chronic hepatitis B
|
|
CR
|
complete response
|
|
ETV
|
entecavir
|
|
HBeAg
|
hepatitis B e antigen
|
|
HBsAg
|
hepatitis B surface antigen
|
|
HBV
|
hepatitis B virus
|
|
LAM
|
lamivudine
|
|
LdT
|
telbivudine
|
|
miRNA
|
microRNA
|
|
NA
|
nucleos(t) ide analogue
|
|
NPV
|
negative predictive value
|
|
OR
|
odds ratio
|
|
peg-IFN
|
pegylated interferon
|
|
PPV
|
positive predictive value
|
|
TDF
|
tenofovir disoproxil fumarate
|
|
ULN
|
upper limit of normal
|
References
|
1
|
Lampertico P, Agarwal K, Berg T, Buti M,
Janssen H, Papatheodoridis G, Zoulim F and Tacke F: EASL 2017
clinical practice guidelines on the management of hepatitis B virus
infection. J Hepatol. 67:370–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Terrault NA, Bzowej NH, Chang KM, Hwang
JP, Jonas MM and Murad MH: American Association for the Study of
Liver Diseases: AASLD guidelines for treatment of chronic hepatitis
B. Hepatology. 63:261–83. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu
Y, Hao Q, Liu Y, Gong H, Zhu Y, et al: A liver-specific microRNA
binds to a highly conserved RNA sequence of hepatitis B virus and
negatively regulates viral gene expression and replication. Faseb
J. 25:4511–4521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen
L, Wu E, Ye X, Gao GF, Wang F, et al: Loss of microRNA 122
expression in patients with hepatitis B enhances hepatitis B virus
replication through cyclin G(1)-modulated P53 activity. Hepatology.
55:730–741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo H, Liu H, Mitchelson K, Rao H, Luo M,
Xie L, Sun Y, Zhang L, Lu Y, Liu R, et al: MicroRNAs-372/373
promote the expression of hepatitis B virus through the targeting
of nuclear factor I/B. Hepatology. 54:808–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X
and Wu Z: miR-101 is down-regulated by the hepatitis B virus ×
protein and induces aberrant DNA methylation by targeting DNA
methyltransferase 3A. Cell Signal. 25:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li F, Zhou P, Deng W, Wang J, Mao R, Zhang
Y, Li J, Yu J, Yang F, Huang Y, et al: Serum microRNA-125b
correlates with hepatitis B viral replication and liver
necroinflammation. Clin Microbiol Infect Dis. 22:384.e1–384.e10.
2016. View Article : Google Scholar
|
|
9
|
Giray BG, Emekdas G, Tezcan S, Ulger M,
Serin MS, Sezgin O, Altintas E and Tiftik EN: Profiles of serum
microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for
HBV-positive hepatocellular carcinoma. Mol Biol Rep. 41:4513–4519.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Chen J, He Y, Zhan X, Zhao R,
Huang Y, Xu H, Zhu Z and Liu Q: miR-125b inhibits hepatitis B virus
expression in vitro through targeting of the SCNN1A gene. Arch
virol. 159:3335–3343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ninomiya M, Kondo Y, Kimura O, Funayama R,
Nagashima T, Kogure T, Morosawa T, Tanaka Y, Nakayama K and
Shimosegawa T: The expression of miR-125b-5p is increased in the
serum of patients with chronic hepatitis B infection and inhibits
the detection of hepatitis B virus surface antigen. J Viral Hepat.
23:330–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liaw YF: Hepatitis flares and hepatitis B
e antigen seroconversion: Implication in anti-hepatitis B virus
therapy. J Gastroenterol Hepatol. 18:246–252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kwon JH, Jang JW, Lee S, Lee J, Chung KW,
Lee YS and Choi J: Pretreatment HBeAg level and an early decrease
in HBeAg level predict virologic response to entecavir treatment
for HBeAg-positive chronic hepatitis B. J Viral Hepat. 19:e41–e47.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kao JH, Asselah T, Dou XG and Hamed K:
Telbivudine therapy for chronic hepatitis B: A journey to identify
super-responders and to optimize treatment using the roadmap model.
J Gastroenterol Hepatol. 32:73–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Lin SM, Ye F, Chen TY, Liu M,
Chen YR, Zheng SQ, Zhao YR and Zhang SL: An early decrease in serum
HBeAg titre is a strong predictor of virological response to
entecavir in HBeAg-positive patients. J Viral Hepat. 18:e184–e190.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan R, Sun J, Yuan Q, Xie Q, Bai X, Ning
Q, Cheng J, Yu Y, Niu J, Shi G, et al: Baseline quantitative
hepatitis B core antibody titre alone strongly predicts HBeAg
seroconversion across chronic hepatitis B patients treated with
peginterferon or nucleos(t)ide analogues. Gut. 65:313–320. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martinot-Peignoux M, Asselah T and
Marcellin P: HBsAg quantification to optimize treatment monitoring
in chronic hepatitis B patients. Liver Int. 35 Suppl:S82–S90. 2015.
View Article : Google Scholar
|
|
18
|
Zoulim F, Carosi G, Greenbloom S, Mazur W,
Nguyen T, Jeffers L, Brunetto M, Yu S and Llamoso C: Quantification
of HBsAg in nucleos(t)ide-naive patients treated for chronic
hepatitis B with entecavir with or without tenofovir in the BE-LOW
study. J Hepatol. 62:56–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu XY, Tan DM, Liu CM, Gu B, Hu LH, Peng
ZT, Chen B, Xie YL, Gong HY, Hu XX, et al: Early hepatitis B viral
DNA clearance predicts treatment response at week 96. World J
Gastroenterol. 23:2978–2986. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu CW, Chao YC, Lee CM, Chang TT and Chen
YC: Efficacy of telbivudine in Taiwanese chronic hepatitis B
patients compared with GLOBE extension study and predicting
treatment outcome by HBV DNA kinetics at week 24. BMC
gastroenterology. 12:1782012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma SW, Huang X, Li YY, Tang LB, Sun XF,
Jiang XT, Zhang YX, Sun J, Liu ZH, Abbott WG, et al: High serum
IL-21 levels after 12 weeks of antiviral therapy predict HBeAg
seroconversion in chronic hepatitis B. J Hepatol. 56:775–781. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo R, Mao H, Hu X, Zheng N, Yan D, He J
and Yang J: Slow reduction of IP-10 Levels predicts HBeAg
seroconversion in chronic hepatitis B patients with 5 years of
entecavir treatment. Sci Rep. 6:370152016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akamatsu S, Hayes CN, Tsuge M, Miki D,
Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, et al:
Differences in serum microRNA profiles in hepatitis B and C virus
infection. J Infect. 70:273–287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alpini G, Glaser SS, Zhang JP, Francis H,
Han Y, Gong J, Stokes A, Francis T, Hughart N, Hubbleet L, et al:
Regulation of placenta growth factor by microRNA-125b in
hepatocellular cancer. J Hepatol. 55:1339–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b
suppressesed human liver cancer cell proliferation and metastasis
by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST,
Nan X, Cao N, Fu CJ, Yan XL, Jia YL, et al: MicroRNA-125b
attenuates epithelial-mesenchymal transitions and targets stem-like
liver cancer cells through small mothers against decapentaplegic 2
and 4. Hepatology. 62:801–815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ,
Kim MG, Chang YG, Shen Q, Park WS, Lee JY, et al: Sirtuin7
oncogenic potential in human hepatocellular carcinoma and its
regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
Hepatology. 57:1055–1067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan DN, Tsang FH, Tam AH, Au SL, Wong CC,
Wei L, Lee JM, He X, Ng IO and Wong CM: Histone lysine
methyltransferase, suppressor of variegation 3–9 homolog 1,
promotes hepatocellular carcinoma progression and is negatively
regulated by microRNA-125b. Hepatology. 57:637–647. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan Z, Yin Y, Zhou J, Wu L, Xu C and Hou
H: Telbivudine treatment of hepatitis B virus-infected pregnant
women at different gestational stages for the prevention of
mother-to-child transmission: Outcomes of telbivudine treatment
during pregnancy. Medicine (Baltimore). 95:e48472016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang W, Wang J, Dang S and Zhuang G:
Cost-effectiveness of antiviral therapy during late pregnancy to
prevent perinatal transmission of hepatitis B virus. Peer J.
4:e17092016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gordon SC, Krastev Z, Horban A, Petersen
J, Sperl J, Dinh P, Martins EB, Yee LJ, Flaherty JF, Kitrinos KM,
et al: Efficacy of tenofovir disoproxil fumarate at 240 weeks in
patients with chronic hepatitis B with high baseline viral load.
Hepatology. 58:505–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hou J, Yin YK, Xu D, Tan D, Niu J, Zhou X,
Wang Y, Zhu L, He Y, Ren H, et al: Telbivudine versus lamivudine in
Chinese patients with chronic hepatitis B: Results at 1 year of a
randomized, double-blind trial. Hepatology. 47:447–454. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Banerjee S, Gunda P, Drake RF and Hamed K:
Telbivudine for the treatment of chronic hepatitis B in
HBeAg-positive patients in China: A health economic analysis.
Springerplus. 5:17192016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Hu P, Qi X, Ren H, Mao RC and
Zhang JM: A comparison of telbivudine and entecavir in the
treatment of hepatitis B e antigen-positive patients: A prospective
cohort study in China. Clin Microbiol Infect. 22(287): e1–9.
2016.PubMed/NCBI
|
|
36
|
Liaw YF, Gane E, Leung N, Zeuzem S, Wang
Y, Lai CL, Heathcote EJ, Manns M, Bzowej N, Niu J, et al: 2-Year
GLOBE trial results: Telbivudine is superior to lamivudine in
patients with chronic hepatitis B. Gastroenterology. 136:486–495.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Z, Huang Y, Wen S, Zhou B and Hou J:
Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res.
37:S36–S41. 2007. View Article : Google Scholar : PubMed/NCBI
|