Introduction
Gastric cancer, one of the most common types of
gastrointestinal cancer in China and a major cause of
cancer-related mortality worldwide, is a serious threat to health
(1–3). Unhealthy eating habits, smoking,
alcohol, viruses and bacteria are risk factors for gastric cancer
(4). Due to poor diagnosis, many
patients are diagnosed with advanced gastric cancer characterized
by extensive invasion and lymphatic metastasis (5,6).
Comprehensive treatment with surgery, chemotherapy and radiotherapy
remains the only effective therapy for gastric cancer (7). Although many improvements have been
made in the treatment of gastric cancer, the overall survival of
patients with gastric cancer is very low, and the prognosis for
patients is not so optimistic (8).
Currently, the traditional drug therapy for tumors generally refers
to chemotherapy; however, normal and malignant cells are destroyed
by chemotherapy, leading to great toxicity to patients (9). Molecular targeted therapy has limited
or nonexistent side effects on normal cells of the body (10). Thus, it is of great importance to
identify novel and effective therapeutic targets for gastric
cancer.
MicroRNA (miRNA), a group of small non-coding RNA,
are able to regulate the expression of downstream targets by
binding to the 3′-untranslated region of the target (UTR) genes
(11,12). Evidence has strongly indicated that
miRNA serve essential roles in the regulation of a variety of
biological progresses, including cell proliferation, apoptosis,
invasion and migration (13).
Additionally, various studies have demonstrated that miRNA have an
important role in gastric cancer progression (14–17).
Upregulator of cell proliferation
(URGCP)/upregulated gene 4 (URG4) is upregulated in a variety of
human cancer types, including hepatocellular carcinoma, epithelial
ovarian cancer, osteosarcoma and gastric cancer (18). URGCP is a tumor promoter, which has
been reported to serve critical roles in gastric cancer cell
proliferation (19–22). TargetScan and MiRanda database
results have suggested that the 3′UTR of URGCP contains the
complementary sequence of miR-671-5p (23).
Previous research has demonstrated that miR-671 was
significantly downregulated in gastric cancer cells compared with
normal gastric cells (24).
Bioinformatic analyses have suggested that the predicted targets of
URGCP were associated with the cell cycle, cell adhesion,
apoptosis, transcription and gene expression (25). As the precise expression and roles of
miR-671-5p in gastric cancer have not been fully elucidated, the
present study aimed to investigate the roles of miR-671-5p in
gastric cancer and explore its mechanism.
Materials and methods
Tissues and cell lines
Fresh gastric cancer tissues and its corresponding
para-carcinoma tissues were obtained from Wujin Hospital affiliated
to Jiangsu University (Changzhou, China). The tissues were provided
by 30 patients with gastric cancer (mean age, 51.2 years; 19 males
and 11 females) between January 2014 and January 2016. The present
study was approved the Ethics Committee of Wujin Hospital
affiliated to Jiangsu University. Patients provided written
informed consent prior to initiation of the study. According to the
seventh edition of TNM classification for gastric cancer (26), 2 (6.7%) patients were Ia stage, 3
(10%) patients were IIa stage, 3 (10%) patients were IIb stage, 10
(33.3%) were IIIa and 12 (40%) were stage IIIc. All tissues were
immediately snap-frozen in liquid nitrogen following surgery.
Gastric cancer cells (MKN28), normal gastric cells
(HFE145) and 293T cells were purchased from Shanghai Bogoo
Biotechnology Co., Ltd., (Shanghai, China). According to research,
the MKN28 cell line is a derivative of the MKN74 gastric tubular
adenocarcinoma cell line (27).
MKN28 cells were routinely grown in RPMI-1640 medium supplemented
with 10% fetal bovine serum (both, Shanghai BioSun Science &
Technology Co., Ltd., Shanghai, China), 1% penicillin and
streptomycin combination at 37°C in a humidified atmosphere with 5%
CO2. HFE145 and 293T cells were cultured in Dulbecco's
modified Eagle's medium supplemented with 10% fetal bovine serum
(Shanghai BioSun Science & Technology Co., Ltd.) at 37°C with
5% CO2.
miR-671-5p target prediction
TargetScan (targetscan.org/vert_71) and MiRanda databases
(mirbase.org) were employed for miR-671-5p target
prediction.
Cell transfection
A total of 24 h prior to transfection, MKN28 cells
(9.5×105/well) were seeded in a 6-well plate. Following
this, the cells were transfected with miR-671-5p mimic (2.5 µg) or
mimic control (cat. no. HMC0002; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) by using Lipofectamine 2000 reagent (10 µl;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
following the manufacturer's protocol. The sequence of the
miR-671-5p mimic used was 5′-AGGAAGCCCUGGAGGGGCUGGAG-3′ (cat. no.
HMI0901; Sigma-Aldrich; Merck KGaA). The plasmids were purchased
from Shanghai Kaiyang Biotechnology Co., Ltd., (Shanghai, China). A
total of 24 h after transfection, cell samples were collected for
further analysis.
Cell proliferation assay
MKN28 cell proliferation ability was detected using
a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) according to the manufacturer's protocol. At 24 h
post-transfection with miR-671-5p mimic or mimic control, MKN28
cells were seeded into a 96-well plate (3×103
cells/well). Subsequently, 20 µl CCK-8 (5 mg/ml) was added to each
well and incubated at 37°C for 4 h. A spectrophotometer was
utilized to determine the optical densities (OD) at 490 nm at 24,
48 and 72 h, respectively. The proliferation rate was determined at
72 h after transfection. All experiments were performed in
triplicate.
Apoptosis analysis assay
To detect cell apoptosis levels, a fluorescein
isothiocyanate (FITC) apoptosis detection kit (Vazyme, Piscataway,
NJ, USA) was utilized, following the manufacturer's protocol. A
total of 24 h after transfection, MKN28 cells were washed three
times with cold PBS solution. Subsequently, MKN28 cells were
re-suspended in 1X binding buffer solution, stained with annexin
V-FITC and propidium iodide, and then incubated for 15 min at room
temperature in the dark. A flow cytometer (BD Biosciences, Franklin
Lakes, NJ, USA) was used to detect the cell apoptotic rate using
WinMDI version 2.5 (Purdue University Cytometry Laboratories, West
Lafayette, IN, USA). Tests were repeated three times.
Western blot analysis
Western blot analysis was performed for protein
expression detection according to standard procedures. The
collected cells were lysed by a radioimmunoprecipitation buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology, Haimen,
China) and then centrifuged at 11,180 × g for 15 min at 4°C. The
protein concentration was determined using a BCA kit. Protein
samples (2 µg/lane) were electrophoresed by 12% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA), then blocked in 5% skimmed milk
(cat. no. 232100; BD Biosciences) for 1 h at room temperature.
Subsequently, membranes were blotted for 1 h at room temperature
with antibodies directed against the following: URGCP (cat. no.
HPA029468; 1.200; Sigma-Aldrich; Merck KGaA), GAPDH (cat. no.
D16H11; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA), B-cell lymphoma 2 (Bcl-2; cat. no. KF364; 1:500; Nanjing
Jiancheng Bioengineering Institute, Jiangsu, China) and
Bcl-2-associated X protein (Bax; cat. no. PR-1381; 1:500; Jiangsu
Sinogene Biotechnology Co., Ltd., Jiangsu, China) Following this,
the membranes were incubated with horseradish peroxidase-labeled
secondary antibodies (cat. no. 58802; 1:1,000; Cell Signaling
Technology, Inc.) for 45 min at room temperature. The membranes
were stained using an electrochemiluminescence kit (cat. no. 32209;
Suzhou Biotsith Bioscience Co., Ltd., Suzhou, China) according to
the manufacturer's protocol. Protein brands were then analyzed by
ImageJ version 1.49 software (National Institute of Health,
Bethesda, MD, USA).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cultured cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. RT reactions were performed
to synthesize cDNA by using a TaqMan miRNA RT kit (cat. no.
4366596FG; Shbybio Co., Ltd., Shanghai, China) according to
manufacturer's protocol. qPCR was subsequently performed using a
QuantiTect SYBR-Green PCR kit (Qiagen, Inc., Valencia, CA, USA), in
line with the manufacturer's protocol. U6 (for miRNA) was used as
an internal control. The thermocycling conditions were 96°C for 5
min (pre-denaturation), 95°C for 30 sec (initiation), 60°C for 30
sec (annealing) and 76°C for 1.5 min (elongation), for a total of
32 cycles. Following this the samples were stored at 4°C. The
sequences of the primers used were as follows: Bcl-2 forward,
5′-GTCTTCGCTGCGGAGATCAT-3′ and reverse,
3′-CATTCCGATATACGCTGGGAC-5′; Bax forward,
5′-CCCGAGAGGTCTTTTTCCGAG-3′ and reverse,
3′-CCAGCCCATGATGGTTCTGAT-5′; URGCP forward,
5′-GACCTTGCTGCCGACATTTAT-3′ and reverse,
3′-GCAGGAAACTGTCTGAGGAGAG-5′; and U6 forward
5′-AGTAAGCCCTTGCTGTCAGTG-3′ and reverse
3′-CCTGGGTCTGATAATGCTGGG-5′. The MystiCq microRNA qPCR Assay
Primers were used as the primers for miR-651-5p (cat. no.
MIRAP00664-250RXN; Sigma-Aldrich; Merck KGaA). In addition, the
relative expression of miR-671-5p and mRNA was quantified using the
2−ΔΔCq method (28).
Tests were performed in triplicate.
Dual-luciferase reporter assay
Wild-type and mutant 3′UTR of URGCP were amplified
and then cloned into the psiCHECK-2 reporter (cat. no. C8021;
Promega Corporation, Madison, WI, USA). miR-671-5p and
miR-671-5p-URGCP-wild type (WT) 3′UTR or miR-671-5p-URGCP-mutant
(MUT) 3′UTR vectors were co-transfected into 293T cells with 30 µl
Lipofectamine 2000 reagent according to the manufacturer's
protocol. A total of 48 h after transfection, the dual-luciferase
reporter assay system (Promega Corporation) was utilized to measure
the luciferase activity. The luciferase activity was then
normalized to the Renilla luciferase activity, which was
used as the internal control.
Statistical analysis
Data were expressed as the mean ± standard
deviation. All statistical analyses were performed using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA). A Student's t-test was performed to
assess the difference between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Reduced expression of miR-671-5p in
gastric cancer tissues and cells
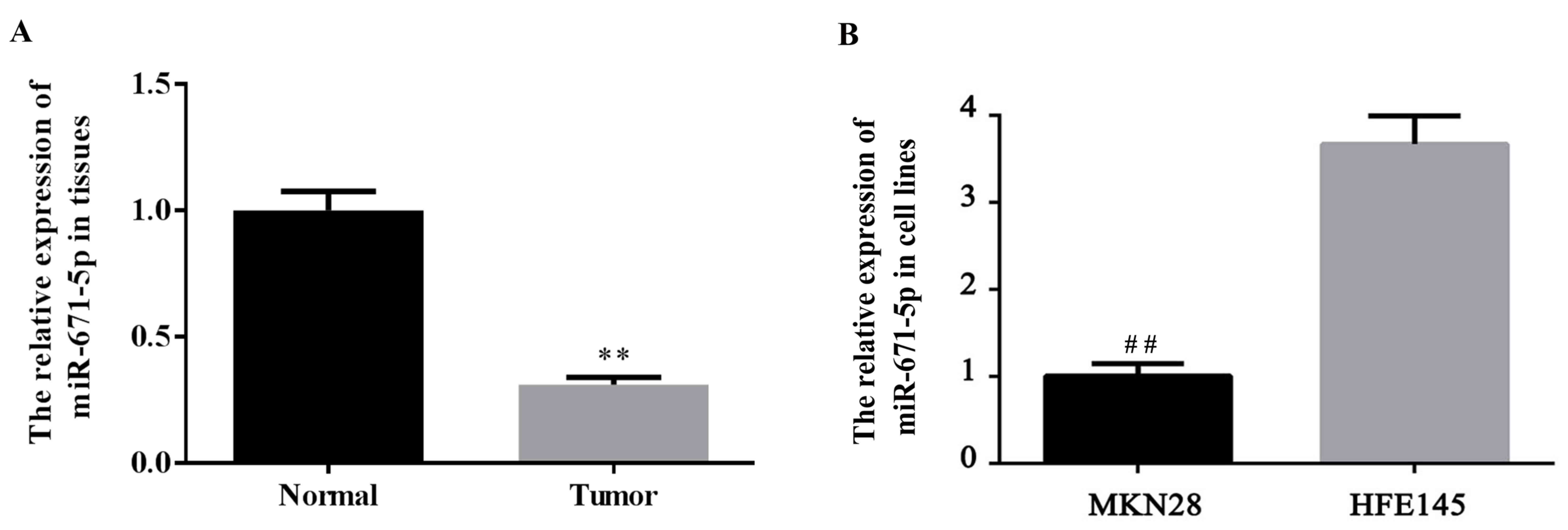
The expression level of miR-671-5p in gastric cancer
tissues and cells was determined. The expression level of
miR-671-5p in tumor and normal gastric tissues, as well as gastric
cancer MKN28 cells and the normal gastric HFE145 cells was detected
using RT-qPCR. As demonstrated in Fig.
1A, the miR-671-5p expression level was significantly decreased
in gastric tumor tissues compared with the levels in para-carcinoma
(normal) tissues (P<0.01). Additionally, as indicated in
Fig. 1B, a significant decrease of
miR-671-5p expression was observed in gastric cancer MKN28 cells
compared with the level in normal gastric HFE145 cells
(P<0.01).
miR-671-5p mimic increases the
expression level of miR-671-5p
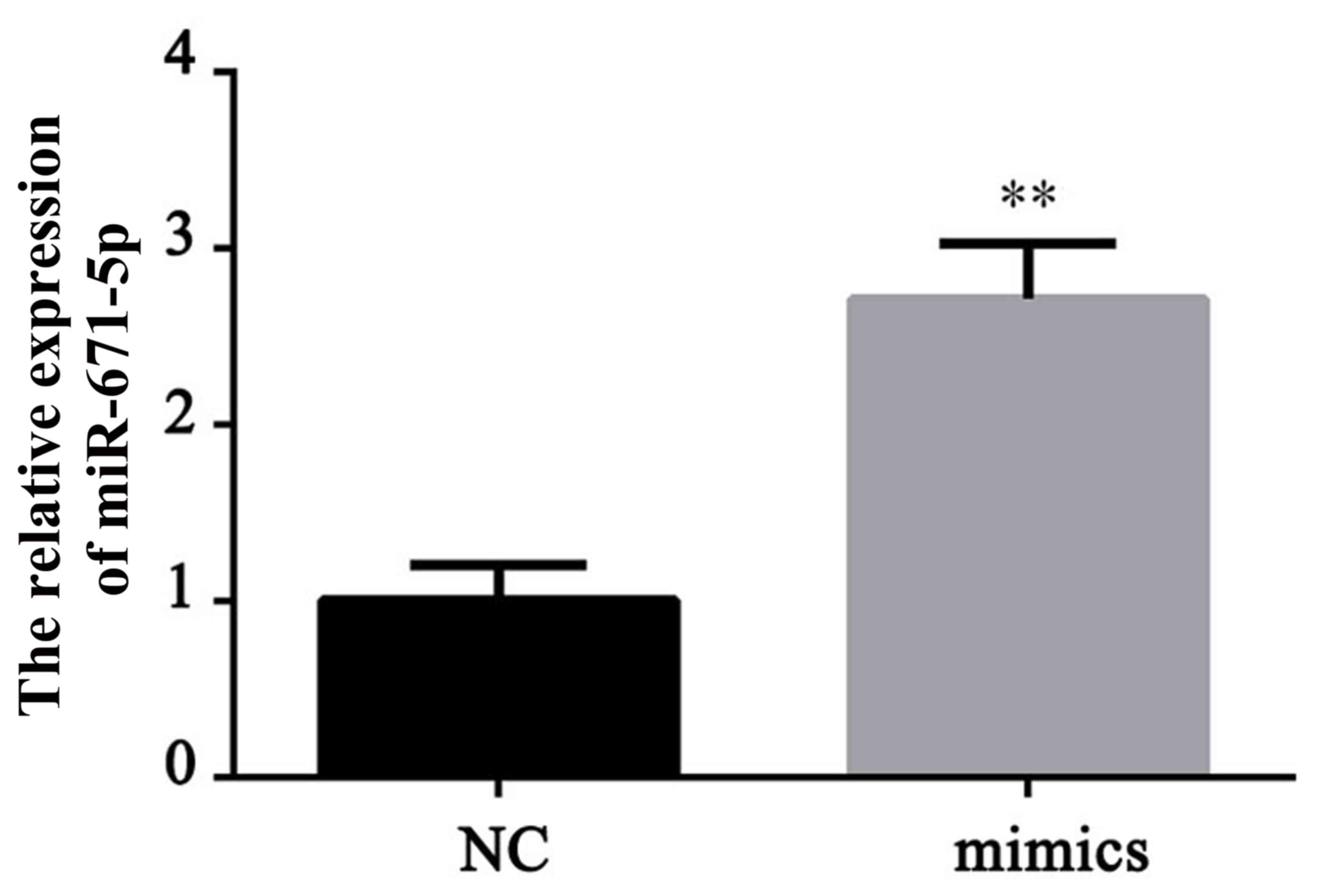
To investigate the role of miR-671-5p in gastric
cancer, a miR-671-5p overexpression cell line was established by
transfection with miR-671-5p mimics. The effective upregulation of
miR-671-5p was demonstrated by RT-qPCR (Fig. 2). The miR-671-5p mimic induced a
significant increase in the expression level of miR-671-5p compared
with the level in the normal control (NC) cells (P<0.01).
miR-671-5p inhibits gastric cancer
cell proliferation
For cell proliferation detection, a CCK-8 kit was
used according to the manufacturer's protocol. A total of 24 h
after MKN28 cells were transfected with miR-671-5p mimics, it was
demonstrated that miR-671-5p mimics significantly suppressed the
proliferation ability of the MKN28 cells compared with the level in
the NC cells, which was indicated by a slower increase of OD value
(P<0.05 at 48 h and P<0.01 at 72 h; Fig. 3). These data indicate that miR-671-5p
inhibits gastric cancer cell proliferation.
miR-671-5p induces gastric cancer cell
apoptosis
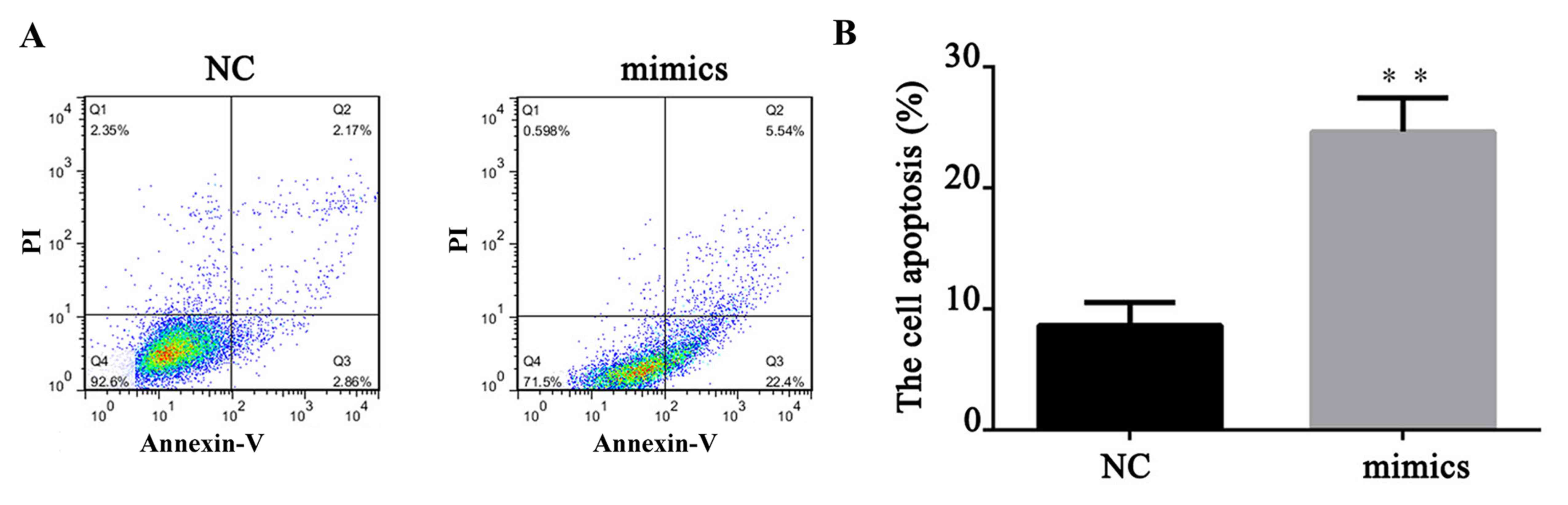
A total of 24 h after transfection, FITC apoptosis
detection was performed to measure the cell apoptosis level. As
demonstrated in Fig. 4, compared
with the NC, upregulation of miR-671-5p significantly increased the
cell apoptosis rate of MKN28 cells (P<0.01). This indicated that
miR-671-5p could induce gastric cancer cell apoptosis. To further
explore the mechanism of the effect of miR-671-5p on cell
apoptosis, the expression levels of cell apoptosis-related proteins
(Bcl-2 and Bax) and corresponding mRNA levels were measured by
western blotting and RT-qPCR. It was demonstrated that the ratio of
Bcl-2/Bax significantly decreased when miR-671-5p was upregulated
in MKN28 cells, with the expression levels of Bcl-2 being
significantly decreased and the levels of Bax being significantly
increased in the mimic group compared with the levels in the NC
group (P<0.01; Fig. 5). Taken
together, these data suggest that miR-671-5p induces gastric cancer
cell apoptosis via regulating the Bcl-2/Bax ratio.
 | Figure 5.miR-671-5p affects the protein
expression of Bcl-2, Bax and URGCP. A total of 24 h after MKN28
cells were transfected with miR-671-5p mimics or its NC, reverse
transcription-quantitative polymerase chain reaction and western
blotting were performed to detect the mRNA and protein expression
levels of Bcl-2, Bax and URGCP. (A) mRNA expression levels of
Bcl-2, Bax and URGCP. (B) Relative protein expression levels of
Bcl-2, Bax and URGCP. (C) Western blotting results of Bcl-2, Bax
and URGCP expression, with GAPDH as the internal reference.
**P<0.01 vs. NC. miR, microRNA; NC, negative control; Bcl-2,
B-cell lymphoma 2; Bax, B-cell lymphoma 2-asssociated X protein;
URGCP, upregulator of cell proliferation. |
URGCP is a direct target of
miR-671-5p
To reveal the mechanism of miR-671-5p function in
gastric cancer, its target gene was predicted using TargetScan and
MiRanda databases (Fig. 6A).
Luciferase reporter gene assay and western blot analysis were
performed to confirm our prediction (Figs. 5C and 6B). The results indicated that the
luciferase activity was significantly declined (P<0.01) in the
293T cells co-transfected with miR-671-5p and miR-671-5p-URGCP-WT,
compared with the NC; however, co-transfection with miR-671-5p and
miR-671-5p-URGCP-MUT did not significantly affect the luciferase
activity level (Fig. 6B).
Additionally, the western blotting results suggested that
miR-671-5p mimics markedly decreased URGCP protein expression
levels compared with the level in the NC cells (Fig. 5C). These data demonstrate that
miR-671-5p inhibits the expression of transcripts containing a
miR-671-5p binding site and that URGCP is a direct target of
miR-671-5p.
Discussion
A large amount of evidence has indicated that miRNA
serve important roles in the development of several types of human
cancer, such as gastric cancer (29,30).
Previous research has revealed that miR-671-5p has critical roles
in human cancer progression. A study by Barbagallo et al
(31) reported that miR-671-5p is
involved in the development of glioblastoma multiform. A study by
Tan et al (32) suggested
that miR-671-5p functions as a tumor suppressor in breast cancer by
targeting forkhead box M1. miR-671-5p has also been demonstrated to
participate in epithelioid sarcoma via the regulation of SMARCB1
expression (33). However, the roles
and underlying mechanisms of miR-671-5p in gastric cancer remain
unclear.
In the present study, it was demonstrated that
miR-671-5p expression was significantly decreased in gastric cancer
MKN28 cells compared with the normal gastric HFE145 cells. The
exact role of miR-671-5p in gastric cancer progression was further
investigated and its underlying mechanisms explored. The present
results suggested that overexpression of miR-671-5p in MKN28 cells
inhibited cell proliferation and induced cell apoptosis. These
results indicated that miR-671-5p functions as a tumor suppressor
in gastric cancer by inhibiting cell proliferation ability and
inducing cell apoptosis.
URGCP/URG4, an upregulator of cell proliferation, is
upregulated in a variety of types of human cancer, including
hepatocellular carcinoma, epithelial ovarian cancer, osteosarcoma
and gastric cancer (19–22). URGCP functions as a tumor promoter,
and it has been reported to serve critical roles in gastric cancer
cell proliferation (22). In the
present study, it was demonstrated that URGCP was a downstream
target of miR-671-5p in human gastric cancer cells. TargetScan and
MiRanda database results suggested that the 3′UTR of URGCP
contained the complementary sequence of miR-671-5p. Subsequently,
the dual-luciferase reporter assay results indicated that
miR-671-5p directly targeted URGCP. Furthermore, overexpression of
miR-671-5p significantly reduced the expression level of URGCP.
Thus, the present study successfully demonstrated that URGCP is a
direct functional target of miR-671-5p in gastric cancer cells.
However, the present study only performed
experiments involving the impact of miR-671-5p on gastric cancer
cells. The pitfall of the present experiment was that the effect of
miR-671-5p on normal gastric cell lines was not studied. Therefore,
future research on the effects of miR-671-5p in normal gastric
cells is required. As miR-671-5p was downregulated in gastric
cancer cells, an miR-671-5p inhibitor should be added to normal
gastric cancer cells to further determine the mechanism of
miR-671-5p in gastric cells.
In summary, the present study demonstrated that
miR-671-5p has a low expression level in gastric cancer cells.
miR-671-5p may serve a role as a tumor suppressor in gastric cancer
by inhibiting cell proliferation and inducing cell apoptosis via
the direct regulation of URGCP. Therefore, the present data
indicated that miR-671-5p may act as a novel therapeutic target in
gastric cancer.
Acknowledgements
The authors would like to thank Wujin Hospital
affiliated to Jiangsu University (Changzhou, China) for its
financial and material support, as well as the supply of
equipment.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guggenheim DE and Shah MA: Gastric cancer
epidemiology and risk factors. J Surg Oncol. 107:230–236. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coburn NG: Lymph nodes and gastric cancer.
J Surg Oncol. 99:199–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi Y and Zhou Y: The role of surgery in
the treatment of gastric cancer. J Surg Oncol. 101:687–692. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kelley JR and Duggan JM: Gastric cancer
epidemiology and risk factors. J Clin Epidemiol. 56:1–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu Q, Sun W, Wang C and Gu Z: Recent
advances of cocktail chemotherapy by combination drug delivery
systems. Adv Drug Deliv Rev. 98:19–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H: Apatinib for molecular targeted
therapy in tumor. Drug Des Devel Ther. 9:6075–6081. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang S, He S, Qiu G, Lu J, Wang J, Liu J,
Fan L, Zhao W and Che X: MicroRNA-125b promotes invasion and
metastasis of gastric cancer by targeting STARD13 and NEU1. Tumour
Biol. 37:12141–12151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu B, Lv X, Su L, Li J, Yu Y, Gu Q, Yan M,
Zhu Z and Liu B: MiR-148a functions as a tumor suppressor by
targeting CCK-BR via inactivating STAT3 and Akt in human gastric
cancer. PLoS One. 11:e01589612016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin G, Zhou H, Xue Y, Yao B and Zhao W:
MicroRNA-340 promotes the tumor growth of human gastric cancer by
inhibiting cyclin G2. Oncol Rep. 36:1111–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cai J, Li R, Xu X, Zhang L, Wu S, Yang T,
Fang L, Wu J, Zhu X, Li M and Huang Y: URGCP promotes non-small
cell lung cancer invasiveness by activating the NF-κB-MMP-9
pathway. Oncotarget. 6:36489–36504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Papp G, Krausz T, Stricker TP, Szendrői M
and Sápi Z: SMARCB1 expression in epithelioid sarcoma is regulated
by miR-206, miR-381, and miR-671-5p on both mRNA and protein
levels. Genes Chromosomes Cancer. 53:168–176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie C, Song LB, Wu JH, Li J, Yun JP, Lai
JM, Xie DY, Lin BL, Yuan YF, Li M and Gao ZL: Upregulator of cell
proliferation predicts poor prognosis in hepatocellular carcinoma
and contributes to hepatocarcinogenesis by downregulating FOXO3a.
PLoS One. 7:e406072012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li WP and Zhou N: URG4 upregulation is
associated with tumor growth and poor survival in epithelial
ovarian cancer. Arch Gynecol Obstet. 286:209–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang J, Zhu B, Lu L, Lian Z, Wang Y, Yang
X, Satiroglu-Tufan NL, Liu J and Luo Z: The expression of novel
gene URG4 in osteosarcoma: Correlation with patients' prognosis.
Pathology. 41:149–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bossel Ben-Moshe N, Avraham R, Kedmi M,
Zeisel A, Yitzhaky A, Yarden Y and Domany E: Context-specific
microRNA analysis: Identification of functional microRNAs and their
mRNA targets. Nucleic Acids Res. 40:10614–10627. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Peng Y, Jin Z, Huang W, Cheng Y,
Liu Y, Feng X, Yang M, Huang Y, Zhao Z, et al: Integrated miRNA
profiling and bioinformatics analyses reveal potential causative
miRNAs in gastric adenocarcinoma. Oncotarget. 6:32878–32889. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song J, Xie H, Lian Z, Yang G, Du R, Du Y,
Zou X, Jin H, Gao J, Liu J and Fan D: Enhanced cell survival of
gastric cancer cells by a novel gene URG4. Neoplasia. 8:995–1002.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marano L, Boccardi V, Braccio B, Esposito
G, Grassia M, Petrillo M, Pezzella M, Porfidia R, Reda G, Romano A,
et al: Comparison of the 6th and 7th editions of the AJCC/UICC TNM
staging system for gastric cancer focusing on the ‘N’
parameter-related survival: The monoinstitutional NodUs Italian
study. World J Surg Oncol. 13:2152015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Capes-Davis A, Theodosopoulos G, Atkin I,
Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA,
Reddel RR and Freshney RI: Check your cultures A list of
cross-contaminated or misidentified cell lines. Int J Cancer.
127:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Petrocca F, Pilozzi E, Rapazzotti M,
Aurello P, Mercantini P, Volinia S, Ruco L, Croce CM and Vecchione
A: MicroRNAs deregulation in gastric cancer. Cancer Res.
66:13382006.
|
|
31
|
Barbagallo D, Condorelli A, Ragusa M,
Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà
A, Battaglia R, et al: Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1
axis is involved in glioblastoma multiforme. Oncotarget.
7:4746–4759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan X, Fu Y, Chen L, Lee W, Lai Y, Rezaei
K, Tabbara S, Latham P, Teal CB, Man YG, et al: miR-671-5p inhibits
epithelial-to-mesenchymal transition by downregulating FOXM1
expression in breast cancer. Oncotarget. 7:293–307. 2016.PubMed/NCBI
|
|
33
|
Song J, Xie H, Lian Z, Yang G, Du R, Du Y,
Zou X, Jin H, Gao J, Liu J and Fan D: Enhanced cell survival of
gastric cancer cells by a novel gene URG4. Neoplasia. 8:995–1002.
2006. View Article : Google Scholar : PubMed/NCBI
|