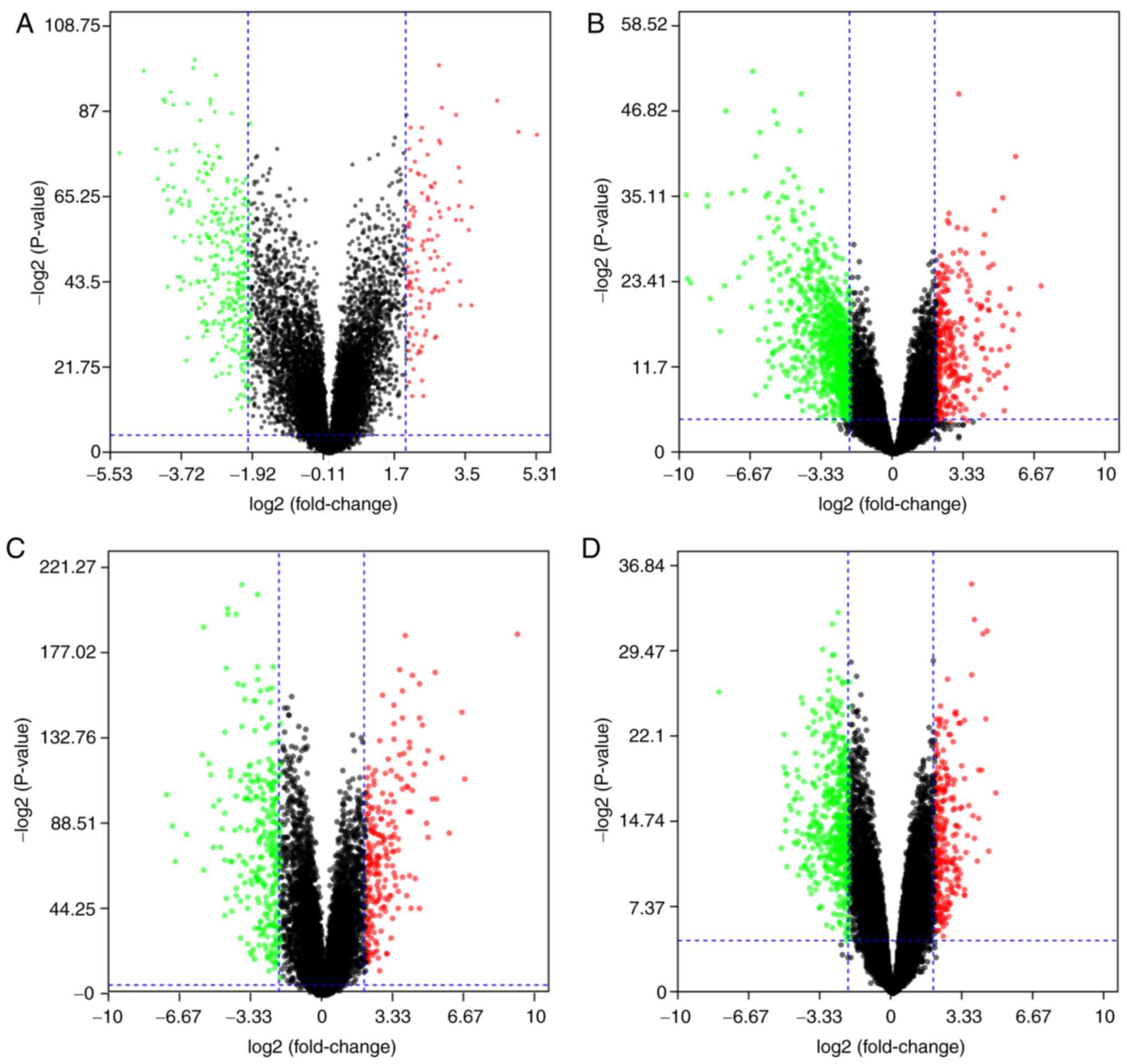
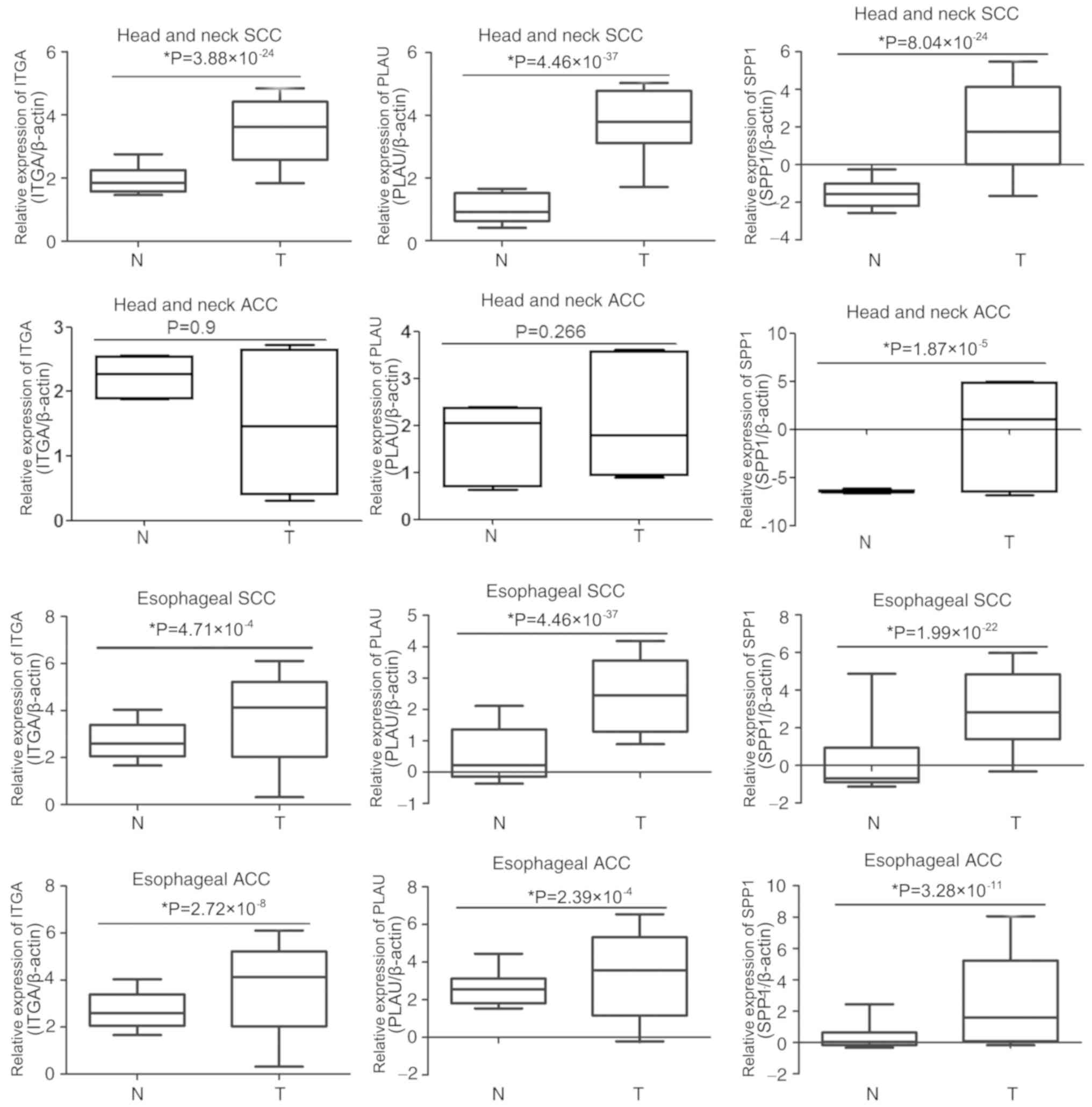
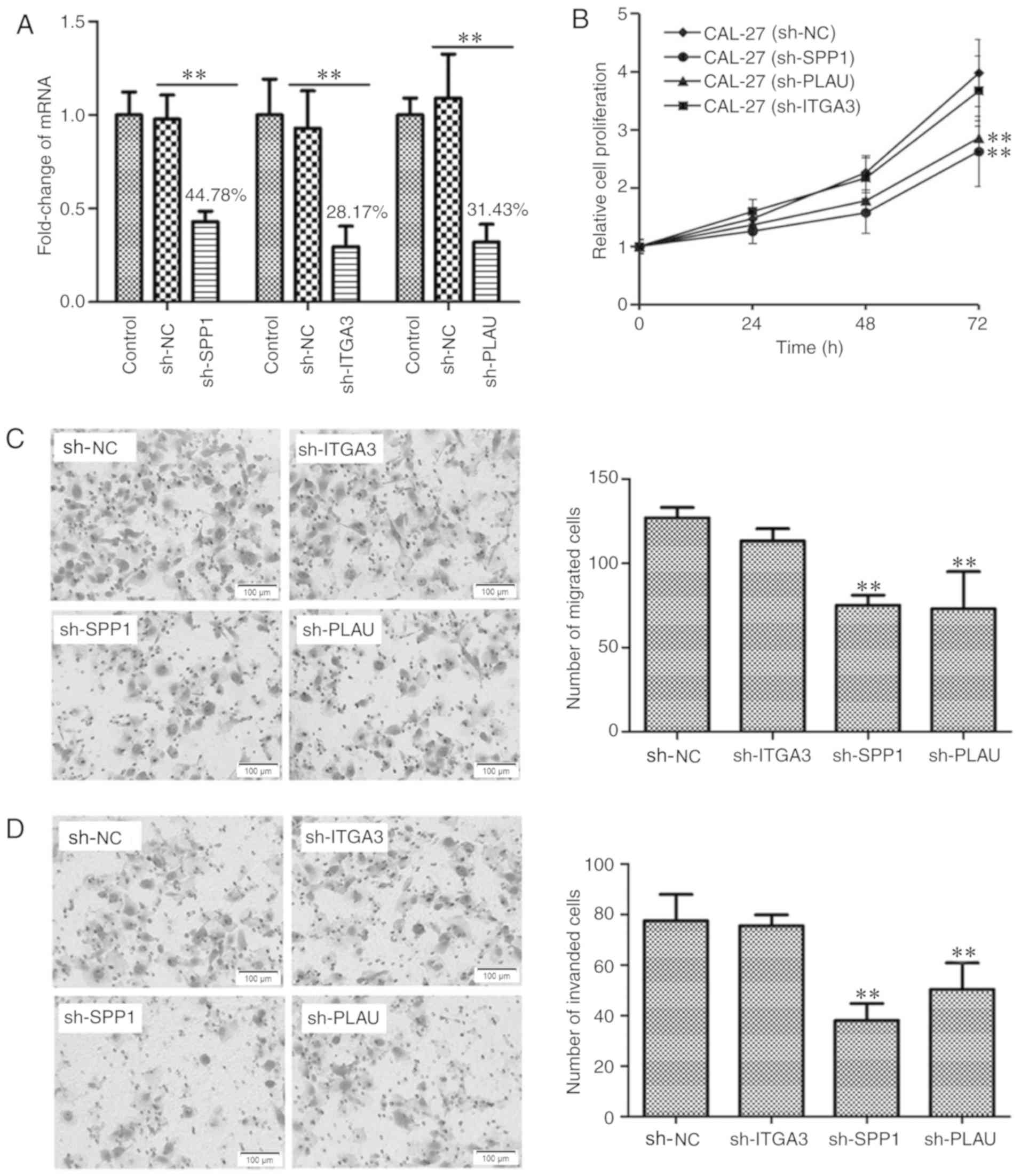
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Petryszak R, Burdett T, Fiorelli B,
Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M,
Kryvych N, et al: Expression atlas update-a database of gene and
transcript expression from microarray- and sequencing-based
functional genomics experiments. Nucleic Acids Res 42 (Database
Issue). D926–D932. 2014. View Article : Google Scholar
|
|
5
|
Randhawa V and Acharya V: Integrated
network analysis and logistic regression modeling identify
stage-specific genes in oral squamous cell carcinoma. BMC Med
Genomics. 8:392015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
7
|
Wang Y, Fan H and Zheng L: Biological
information analysis of differentially expressed genes in oral
squamous cell carcinoma tissues in GEO database. J BUON.
23:1662–1670. 2018.PubMed/NCBI
|
|
8
|
Mitchell KA, Zingone A, Toulabi L,
Boeckelman J and Ryan BM: Comparative transcriptome profiling
reveals coding and noncoding RNA differences in NSCLC from African
Americans and European Americans. Clin Cancer Res. 23:7412–7425.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hardiman G, Savage SJ, Hazard ES, Wilson
RC, Courtney SM, Smith MT, Hollis BW, Halbert CH and Gattoni-Celli
S: Systems analysis of the prostate transcriptome in
African-American men compared with European-American men.
Pharmacogenomics. 17:1129–1143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee CH, Chang JS, Syu SH, Wong TS, Chan
JY, Tang YC, Yang ZP, Yang WC, Chen CT, Lu SC, et al: IL-1beta
promotes malignant transformation and tumor aggressiveness in oral
cancer. J Cell Physiol. 230:875–884. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambatipudi S, Gerstung M, Pandey M, Samant
T, Patil A, Kane S, Desai RS, Schäffer AA, Beerenwinkel N and
Mahimkar MB: Genome-wide expression and copy number analysis
identifies driver genes in gingivobuccal cancers. Genes Chromosomes
Cancer. 51:161–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Méndez E, Houck J, Fan W,
Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG,
et al: Gene expression profiling identifies genes predictive of
oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
17:2152–2162. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res 41
(Database Issue). D808–D815. 2013.
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Zhu R, Huang Z, Li H and Zhu H:
Lipopolysaccharide-induced toll-like receptor 4 signaling in cancer
cells promotes cell survival and proliferation in hepatocellular
carcinoma. Dig Dis Sci. 58:2223–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koundouros N and Poulogiannis G:
Phosphoinositide 3-kinase/Akt signaling and redox metabolism in
cancer. Front Oncol. 8:1602018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gay NJ, Symmons MF, Gangloff M and Bryant
CE: Assembly and localization of Toll-like receptor signalling
complexes. Nat Rev Immunol. 14:546–558. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shcheblyakov DV, Logunov DY, Tukhvatulin
AI, Shmarov MM, Naroditsky BS and Gintsburg AL: Toll-like receptors
(TLRs): The role in tumor progression. Acta Naturae. 2:21–29.
2010.PubMed/NCBI
|
|
21
|
Wang W and Wang J: Toll-like receptor 4
(TLR4)/Cyclooxygenase-2 (COX-2) regulates prostate cancer cell
proliferation, migration, and invasion by NF-kappaB activation. Med
Sci Monit. 24:5588–5597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang F, Jin R, Zou BB, Li L, Cheng FW, Luo
X, Geng X and Zhang SQ: Activation of toll-like receptor 7
regulates the expression of IFN-λ1, p53, PTEN, VEGF, TIMP-1 and
MMP-9 in pancreatic cancer cells. Mol Med Rep. 13:1807–1812. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin CH, Lin HH, Kuo CY and Kao SH:
Aeroallergen Der p 2 promotes motility of human non-small cell lung
cancer cells via toll-like receptor-mediated up-regulation of
urokinase-type plasminogen activator and integrin/focal adhesion
kinase signaling. Oncotarget. 8:11316–11328. 2017.PubMed/NCBI
|
|
24
|
Braunstein MJ, Kucharczyk J and Adams S:
Targeting toll-like receptors for cancer therapy. Target Oncol.
13:583–598. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yeh DW, Chen YS, Lai CY, Liu YL, Lu CH, Lo
JF, Chen L, Hsu LC, Luo Y, Xiang R and Chuang TH: Downregulation of
COMMD1 by miR-205 promotes a positive feedback loop for amplifying
inflammatory- and stemness-associated properties of cancer cells.
Cell Death Differ. 23:841–852. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tokunaga R, Zhang W, Naseem M, Puccini A,
Berger MD, Soni S, McSkane M, Baba H and Lenz HJ: CXCL9, CXCL10,
CXCL11/CXCR3 axis for immune activation - A target for novel cancer
therapy. Cancer Treat Rev. 63:40–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan S, Wang K, Sun F, Li Y and Gao Y:
CXCL9 promotes prostate cancer progression through inhibition of
cytokines from T cells. Mol Med Rep. 18:1305–1310. 2018.PubMed/NCBI
|
|
28
|
Bronger H, Singer J, Windmüller C, Reuning
U, Zech D, Delbridge C, Dorn J, Kiechle M, Schmalfeldt B, Schmitt M
and Avril S: CXCL9 and CXCL10 predict survival and are regulated by
cyclooxygenase inhibition in advanced serous ovarian cancer. Br J
Cancer. 115:553–563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bronger H, Karge A, Dreyer T, Zech D,
Kraeft S, Avril S, Kiechle M and Schmitt M: Induction of cathepsin
B by the CXCR3 chemokines CXCL9 and CXCL10 in human breast cancer
cells. Oncol Lett. 13:4224–4230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Spaks A: Role of CXC group chemokines in
lung cancer development and progression. J Thorac Dis. 9 (Suppl
3):S164–S171. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang C, Li Z, Xu L, Che X, Wen T, Fan Y,
Li C, Wang S, Cheng Y, Wang X, et al: CXCL9/10/11, a regulator of
PD-L1 expression in gastric cancer. BMC Cancer. 18:4622018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ruiduo C, Ying D and Qiwei W: CXCL9
promotes the progression of diffuse large B-cell lymphoma through
up-regulating beta-catenin. Biomed Pharmacother. 107:689–695. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han B, Feng D, Yu X, Liu Y, Yang M, Luo F,
Zhou L and Liu F: MicroRNA-144 mediates chronic inflammation and
tumorigenesis in colorectal cancer progression via regulating C-X-C
motif chemokine ligand 11. Exp Ther Med. 16:1935–1943.
2018.PubMed/NCBI
|
|
34
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Spangle JM, Roberts TM and Zhao JJ: The
emerging role of PI3K/AKT-mediated epigenetic regulation in cancer.
Biochim Biophys Acta Rev Cancer. 1868:123–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao Y, Xia F, Wang P and Gao M:
MicroRNA935p promotes the progression of human retinoblastoma by
regulating the PTEN/PI3K/AKT signaling pathway. Mol Med Rep.
18:5807–5814. 2018.PubMed/NCBI
|
|
37
|
Lunardi A, Webster KA, Papa A, Padmani B,
Clohessy JG, Bronson RT and Pandolfi PP: Role of aberrant PI3K
pathway activation in gallbladder tumorigenesis. Oncotarget.
5:894–900. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dong J, Zhai B, Sun W, Hu F, Cheng H and
Xu J: Activation of phosphatidylinositol 3-kinase/AKT/snail
signaling pathway contributes to epithelial-mesenchymal
transition-induced multi-drug resistance to sorafenib in
hepatocellular carcinoma cells. PLoS One. 12:e01850882017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu S, Li Y, Lu Y, Huang J, Ren J, Zhang S,
Yin Z, Huang K, Wu G and Yang K: LZTS2 inhibits PI3K/AKT activation
and radioresistance in nasopharyngeal carcinoma by interacting with
p85. Cancer Lett. 420:38–48. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu W, Yang Z, Xie C, Zhu Y, Shu X, Zhang
Z, Li N, Chai N, Zhang S, Wu K, et al: PTEN lipid phosphatase
inactivation links the hippo and PI3K/Akt pathways to induce
gastric tumorigenesis. J Exp Clin Cancer Res. 37:1982018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hamzehzadeh L, Atkin SL, Majeed M, Butler
AE and Sahebkar A: The versatile role of curcumin in cancer
prevention and treatment: A focus on PI3K/AKT pathway. J Cell
Physiol. 233:6530–6537. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang D, Du C, Ji D, Xi J and Gu J:
Overexpression of LAMC2 predicts poor prognosis in colorectal
cancer patients and promotes cancer cell proliferation, migration,
and invasion. Tumour Biol. 39:10104283177058492017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Korbakis D, Dimitromanolakis A, Prassas I,
Davis GJ, Barber E, Reckamp KL, Blasutig I and Diamandis EP: Serum
LAMC2 enhances the prognostic value of a multi-parametric panel in
non-small cell lung cancer. Br J Cancer. 113:484–491. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Moon YW, Rao G, Kim JJ, Shim HS, Park KS,
An SS, Kim B, Steeg PS, Sarfaraz S, Changwoo Lee L, et al: LAMC2
enhances the metastatic potential of lung adenocarcinoma. Cell
Death Differ. 22:1341–1352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hubbard NE, Chen QJ, Sickafoose LK, Wood
MB, Gregg JP, Abrahamsson NM, Engelberg JA, Walls JE and Borowsky
AD: Transgenic mammary epithelial osteopontin (spp1) expression
induces proliferation and alveologenesis. Genes Cancer. 4:201–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ng L, Wan T, Chow A, Iyer D, Man J, Chen
G, Yau TC, Lo O, Foo CC, Poon JT, et al: Osteopontin overexpression
induced tumor progression and chemoresistance to oxaliplatin
through induction of stem-like properties in human colorectal
cancer. Stem Cells Int. 2015:2478922015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Choe EK, Yi JW, Chai YJ and Park KJ:
Upregulation of the adipokine genes ADIPOR1 and SPP1 is related to
poor survival outcomes in colorectal cancer. J Surg Oncol.
117:1833–1840. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zeng B, Zhou M, Wu H and Xiong Z: SPP1
promotes ovarian cancer progression via Integrin β1/FAK/AKT
signaling pathway. Onco Targets Therapy. 11:1333–1343. 2018.
View Article : Google Scholar
|
|
49
|
Zhang W, Fan J, Chen Q, Lei C, Qiao B and
Liu Q: SPP1 and AGER as potential prognostic biomarkers for lung
adenocarcinoma. Oncol Lett. 15:7028–7036. 2018.PubMed/NCBI
|
|
50
|
Zhang Y, Du W, Chen Z and Xiang C:
Upregulation of PD-L1 by SPP1 mediates macrophage polarization and
facilitates immune escape in lung adenocarcinoma. Exp Cell Res.
359:449–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
He F, Chen H, Probst-Kepper M, Geffers R,
Eifes S, Del Sol A, Schughart K, Zeng AP and Balling R: PLAU
inferred from a correlation network is critical for suppressor
function of regulatory T cells. Mol Syst Biol. 8:6242012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kurozumi A, Goto Y, Matsushita R, Fukumoto
I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa
T and Seki N: Tumor-suppressive microRNA-223 inhibits cancer cell
migration and invasion by targeting ITGA3/ITGB1 signaling in
prostate cancer. Cancer Sci. 107:84–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sakaguchi T, Yoshino H, Yonemori M,
Miyamoto K, Sugita S, Matsushita R, Itesako T, Tatarano S, Nakagawa
M and Enokida H: Regulation of ITGA3 by the dual-stranded
microRNA-199 family as a potential prognostic marker in bladder
cancer. Br J Cancer. 116:1077–1087. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sa KD, Zhang X, Li XF, Gu ZP, Yang AG,
Zhang R, Li JP and Sun JY: A miR-124/ITGA3 axis contributes to
colorectal cancer metastasis by regulating anoikis susceptibility.
Biochem Biophys Res Commun. 501:758–764. 2018. View Article : Google Scholar : PubMed/NCBI
|