Introduction
Lung cancer has been reported to be the leading
cause of cancer-associated death (1). Non-small cell lung carcinoma (NSCLC),
with a 5-year survival rate of only 11%, accounts for ~80% of cases
of lung cancer (2). The majority of
cases of mortality due to cancer are caused by tumor invasion and
metastasis, which have been acknowledged as the major reasons for
disease progression and therapy failure (3). Therefore, the inhibition of tumor
metastasis may be an important anti-cancer strategy.
MicroRNAs (miRNAs or miRs) are a group of small
non-coding RNAs (18–22 nucleotides), which can regulate the
expression of target mRNAs by binding to their 3′-untranslated
region (UTR) (4). miRNAs are
associated with a variety of cellular functions, including
proliferation, apoptosis, differentiation and migration (5). miRNAs have emerged as important gene
regulatory molecules in various cancers, functioning either as
tumor suppressors or oncogenes; their aberrant expression and
dysregulation are closely associated with the progression of cancer
(6,7). These functional miRNAs may be ideal
diagnostic biomarkers and therapeutic targets for cancers. Among
these functional miRNAs, miR-191 was reported to be notably
upregulated and acted as an oncogenic member in breast (8), gastric (9) and pancreatic cancers (10), hepatocellular carcinoma (HCC)
(11), colorectal carcinoma (CRC)
(12), and intrahepatic
cholangiocarcinoma (ICC) (13). The
oncogenic effects of enhanced miR-191 expression levels are exerted
on a number of cancer-specific signaling pathways. For example, the
signal cascades, including miR-191-Ten-eleven translocation (TET)
methylcytosine dioxygenase 1a-p53 and miR-191-ubiquitin-specific
peptidase 10-p53 were identified as pivotal pathways that promote
the invasion and progression of ICC and pancreatic cancer,
respectively (10,13). Increased miRNA-191 expression levels
induced the invasion and migration of CRC cell lines via the
downregulation of tissue inhibitor of metalloproteinase 3 (TIMP3),
thus upregulating its downstream gene, matrix metalloproteinase 3
(12). TIMP3 was also identified as
a target gene regulated by miR-191 in HCC cell lines (14). Despite advances in understanding the
oncogenic roles of miR-191 in various human cancers, the role of
miR-191 in NSCLC requires further investigation.
CCAAT/enhanced binding protein β (C/EBPβ), a member
of C/EBP family of transcription factors, serves a critical role in
the modulation of cell growth and development (15,16). The
roles of C/EBP in various cancers, including NSCLC, have been
discussed previously (17,18). Some evidence suggests that C/EBPβ may
act as a key factor for cancer development; however, its role is
reportedly controversial: Although C/EBPβ has been observed to be
upregulated and to act as an oncogene in CRC (19), ovarian (20) and prostate cancers (21), as well as glioma (22), it has also been demonstrated to act
as a tumor suppressor by inhibiting tumor migration in breast
cancer (23,24) and HCC (25).
Given the pivotal roles of miR-191 in tumor
progression, it was speculated that miR-191 may be associated with
NSCLC invasion and metastasis. In the present study, the expression
of miR-191 in HSCLC tissues was investigated; the overexpression of
miR-191 was demonstrated to induce the migration and invasion of
the A549 NSCLC cell line. Bioinformatic prediction and luciferase
reporter assays suggested that C/EBPβ was a direct target of
miR-191. The present results indicated that miR-191 may act as a
potent promoter of tumorigenesis in NSCLC possibly through the
negative regulation of C/EBPβ, which may suggest a novel mechanism
and provide a basis for the diagnosis and therapy for NSCLC
patients.
Materials and methods
Patient samples
Tumor samples and their corresponding adjacent
tissues were collected from 30 patients (age range, 32–71 years)
with NSCLC admitted to Anqiu People's Hospital (Anqiu, China)
between April 2016 and March 2017. These clinical samples were
frozen immediately in liquid nitrogen and stored at −80°C until
use. Patients were included in the present study if they had not
received chemotherapy or radiotherapy prior to recruitment. Details
on the patients' characteristics, including gender, age, grade and
stage are presented in Table I.
Written informed consent was obtained from all patients and all
study protocols were approved by the local ethics committee of
Anqiu People's Hospital (Anqiu, China).
 | Table I.Expression of miR-191 and C/EBPβ in
lung carcinoma patient tissues. |
Table I.
Expression of miR-191 and C/EBPβ in
lung carcinoma patient tissues.
| Characteristic | Patients (n) | miR-191 | P-value | C/EBPβ | P-value |
|---|
| Sex |
|
| 0.341 |
| 0.472 |
|
Male | 17 | 1.744±0.094 |
| 0.666±0.039 |
|
|
Female | 13 | 1.605±0.109 |
| 0.627±0.034 |
|
| Age (years) |
|
| 0.188 |
| 0.230 |
|
<60 | 10 | 1.516±0.072 |
| 0.688±0.035 |
|
|
≥60 | 20 | 1.621±0.021 |
| 0.624±0.019 |
|
| Histological
grade |
|
| 0.005 |
| 0.022 |
|
Well-intermediate
differentiation | 18 | 1.659±0.039 |
| 0.597±0.028 |
|
| Poor
differentiation | 12 | 1.503±0.025 |
| 0.692±0.024 |
|
| Metastasis |
|
| 0.013 |
| <0.001 |
| No | 17 | 1.651±0.046 |
| 0.576±0.019 |
|
|
Yes | 13 | 1.470±0.050 |
| 0.704±0.023 |
|
Cell culture
The human NSCLC cell line A549 and human embryonic
kidney cells 293T were purchased from the Shanghai Cell Bank,
Chinese Academy of Sciences (Shanghai, China). Cells were cultured
in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), supplemented with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin
and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.), and incubated at 37°C in a humidified atmosphere of 5%
CO2. Cells were passaged at 90% confluence with 0.25%
Trypsin-EDTA.
miR-191 transfection
Using the transfection reagent Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol, A549 cells were transiently transfected
with 50 nM miR-191 mimic, 50 nM miR-191 mimic negative control, or
200 nM miR-191 inhibitor, 200 nM miR-191 inhibitor NC, 30 nM C/EBPβ
small interfering RNA (siRNA) or C/EBPβ siRNA NC (scramble
sequences) which were designed and synthesized by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). The transfection efficiency was
measured at 72 h post-transfection using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Transfected cells were continuously cultured for subsequent
experiments. The sequences of miR-191 mimics, inhibitors and NCs
were as follows: miR-191 mimics, forward
5′-CAACGGAAUCCCAAAAGCAGCUG-3′ and reverse
5′-GCUGCUUUUGGGAUUCCGUUGUU-3′; NC mimics, forward
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse
5′-ACGUGACACGUUCGGAGAATT-3′; miR-191-5p inhibitor,
5′-CAGCUGCUUUUGGGAUUCCGUUG-3′; NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAA-3′; C/EBPβ siRNA1 forward,
CCCGTGGTGTTATTTAAAGAA and reverse, 5′-UAAGCGAUUACUCAGGGCCCG-3′;
C/EBPβ siRNA2 forward, 5′-AGAACGAGCGGCTGCAGAAGA-3′ and reverse,
5′-AGAGGAATTCCAGTATTAGC-3′; C/EBPβ siRNA NC, forward,
AATTCTCCGAACGTGTCACGT and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′.
RT-qPCR
To detect expression of miR-191, total RNA was
extracted from the tissues of patients with NSCLC or A549 cells
using the mirVana miRNA isolation kit (Ambion; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Total RNA was subsequently transcribed into cDNA using a TaqMan
MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.). RT-qPCR analysis was performed using
TaqMan Advanced miRNA assays (Thermo Fisher Scientific, Inc.)
according to the protocols of the manufacturer under the following
thermocycling conditions: 95°C for 2 min, followed by 30 cycles at
94°C for 45 sec, 55°C for 55 sec, 72°C for 1 min and 72°C for 10
min. To quantify the amount of C/EBPβ mRNA, total RNA was extracted
from the tissue samples of patients with NSCLC or A549 cells at 48
h post-transfection using an RNeasy Mini kit (Qiagen, Inc.,
Valencia, CA, USA), and transcribed into cDNA using a
primeScript® RT reagent kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). A SYBR green qPCR assay kit (Qiagen, Inc.) was
carried out to measure C/EBPβ expression levels. U6 and a-GAPDH
were used as internal controls for miR-191 and C/EBPβ,
respectively. Samples were tested in triplicate, and the
differences in threshold cycles between the target genes and
house-keeping genes (U6 in miRNA and GAPDH in mRNA) were calculated
using the 2−ΔΔCq method (26). The primers used were as follows:
miR-191, forward 5′-AAGGGAATCTTTCTGCACTCAAGCAT-3′ and reverse
5′-ATGCTTGAGTGCAGAAAGATTCCCTT-3′; U6, forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-ACGCTTCACGAATTTGCGT-3′;
C/EBPβ, forward 5′-TTCAAGCAGCTGCCCGAGCC-3′ and reverse,
5′-GCCAAGTGCCCCAGTGCCAA-3′; and GAPDH, forward
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse
5′-GAAGATGGTGATGGGATTTC-3′.
Western blotting
A549 cells were transfected with miR-191 for 48 h,
as detailed above. For the immunoblotting analysis of C/EBPβ, the
cells were lysed in ice-cold lysis buffer containing a protease
inhibitor cocktail (Roche Diagnostics, Basel, Switzerland), and the
protein extracts were denatured in a boiling water bath for 10 min.
The concentration of protein was determined using a bicinchoninic
acid kit (Thermo Fisher Scientific, Inc.). Total protein (30 µg)
was separated by 10% SDS-PAGE and transferred to nitrocellulose
membranes. Following blocking with 5% skimmed milk overnight at
4°C, the membranes were incubated with primary antibodies against
C/EBPβ (ab53138; 1:1,000) and GAPDH (ab9485; 1:1,000; both Abcam,
Cambridge, MA, USA) overnight at 4°C, followed by incubation with
horseradish peroxidase-conjugated secondary antibodies (ab205718;
1:2,000; Abcam) at room temperature for 60 min. The target protein
was detected with a chemiluminescent kit (GE Healthcare Life
Sciences, Little Chalfont, UK). Protein was quantitatively analyzed
using Image J 1.48 u software (National Institutes of Health,
Bethesda, MD, USA).
Scratch-wound healing assay
Transfected A549 cells were cultured in 6-well
plates until 90% confluence was attained. Scratches were formed by
drawing two parallel lines with a 10 µl sterile pipette tip. Any
cellular debris were removed by washing the cells three times with
PBS. The scratched layer was then incubated in fresh media at 37°C.
The area of migration was measured at 0, 12 and 24 h under a light
microscope (magnification, ×200).
Transwell invasion assay
Invasion assays were performed using a Neuro Probe
Standard 24-well Chemotaxis Chamber (pore size, 8 µm; EMD
Millipore, Billerica, MA, USA) following the manufacturer's
protocol. Briefly, the filter inserts were coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). A total of 100 µl of
3×105/ml transfected A549 cells were added into the
inserts (top chamber) with serum-free medium (RPMI 1640), and the
bottom chambers were coated with 1 ml 10% FBS-containing media.
Following 24 h incubation at 37°C, non-migrated cells in the top
chamber were removed and inserts were fixed in methanol at room
temperature for 10 min. The inserts were then stained with crystal
violet staining solution at room temperature for 5 min and the
cells in five randomly selected fields were counted under a light
microscope (Olympus Corporation, Tokyo, Japan; magnification,
×200).
Dual-luciferase reported assay
C/EBPβ was predicted to be a target gene of miR-191
based on analysis with TargetScan (http://www.targetscan.org). The 3′UTR sequence of
C/EBPβ was amplified by PCR from genomic NSCLC cell DNA as
aforementioned and then inserted into the multiple cloning site
downstream of the luciferase reporter gene in the pMIR-REPORT™
luciferase plasmid (Thermo Fisher Scientific, Inc.) to construct
the luciferase reporter plasmid (C/EBPβ 3′UTR wt). To generate the
C/EBPβ 3′UTR mutated reporter (C/EBPβ 3′UTR mut), several
nucleotides in the C/EBPβ 3′UTR that bind the seed region of
miR-191 were mutated via PCR as described previously (26). All constructed plasmids were verified
via DNA sequencing also as described previously (26). C/EBPβ 3′UTR wt or mut, and miR-191 or
NC mimic were transfected into 293T cells using Lipofectamine 2000.
After 24 h, the cells were lysed and their luciferase activities
were measured using a dual-luciferase detection kit (cat. no.
RG027; Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's instructions. The levels of firefly
luciferase were presented as a ratio to Renilla internal
control. Primers for mutant construction were as follows: Forward,
5′-AAGGGAATCTTTCTGCACTCAAGCAT-3′ and reverse
5′-ATGCTTGAGTGCAGAAAGATTCCCTT-3′.
Bioinformatics analysis
The potential targets of miR-191 were predicted by
TargetScan (https://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/). The search term used
was has-miR-191.
Statistical analysis
All results were analyzed using GraphPad Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA) and the
values were presented as the mean ± standard deviation from at
least three independent experiments. Statistical significance was
analyzed using paired or unpaired Student's t-test. One-way
Analysis of variance with Turkey's post hoc test was performed to
analyze the differences among multiple groups. The clinical
information of the patients was examined via χ2 test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
The expression of miR-191 and C/EBPβ
in the tumors of patients with NSCLC
miR-191 has been reported to be highly expressed in
a variety of solid cancers. RT-qPCR analysis was performed to
determine the expression levels of miR-191 in human NSCLC tissues.
It was demonstrated that miR-191 expression levels in tumor tissues
were significantly higher than those in adjacent non-tumor control,
while the expression of C/EBPβ was significantly downregulated
(Fig. 1; P<0.01). The
associations between miR-191 expression and clinical
characteristics were further analyzed (Table I). A significant association was
observed between miR-191 expression, grade and metastasis (P=0.005
and 0.013, respectively). No significant differences were observed
between miR-191 expression and other clinical data, including
gender and age. The results also demonstrated that there may be a
strong association of miR-191 expression and NSCLC progression.
Effect of miR-191 mimics and miR-191
inhibitor on miR-191 expression in A549 cells
To further confirm the roles of miR-191 in NSCLC,
cells were transfected with miR-191 mimics and miR-191 inhibitor
respectively. As presented in Fig.
2A, the expression of miR-191 in the cells transfected with
miR-191 mimics was significantly increased, compared with the cells
transfected with miR-191 NC mimics (P<0.01). However, the
expression levels of miR-191 were decreased following the treatment
of miR-191 inhibitor in comparison with miR-191 NC inhibitor
(Fig. 2B; P<0.01).
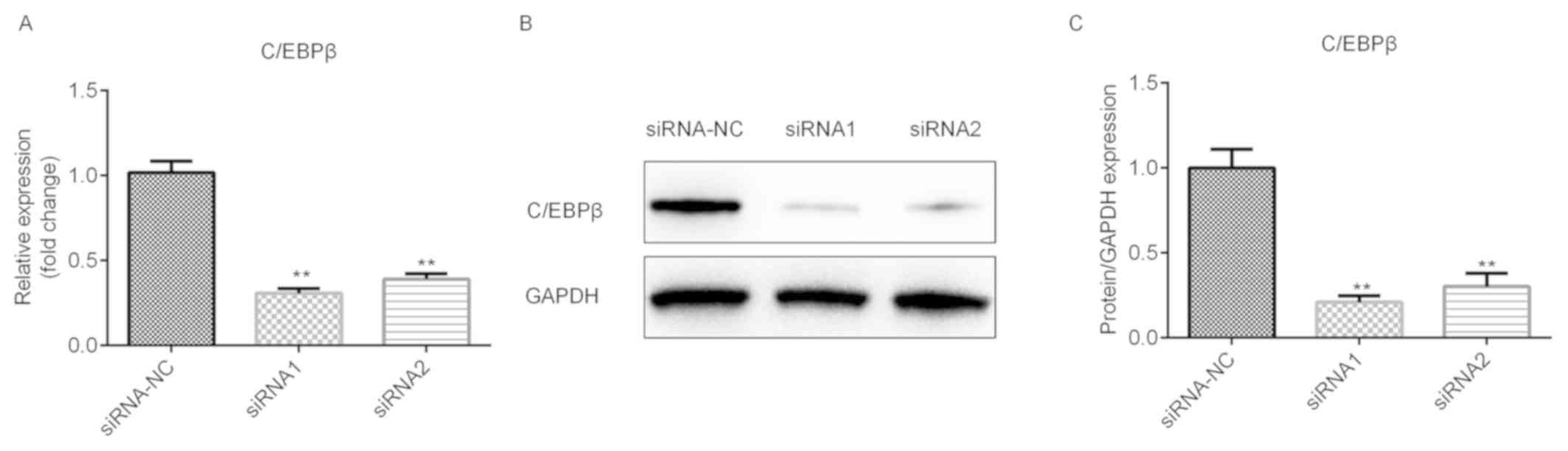
Effects of si-C/EBPβ on the expression
of C/EBPβ in A549 cells
The potential roles of si-C/EBPβ in NSCLC were
further explored. The results demonstrated that the mRNA level of
C/EBPβ was significantly downregulated following the transfection
of siRNA1 and siRNA2, and the siRNA1 was more potent (Fig. 3A; P<0.01), which is in accordance
with the protein level (Fig. 3B and
C; P<0.01).
miR-191 promotes NSCLC cell migration
and invasion in vitro
The effects of miR-191 on the migration and invasion
of A549 cells were evaluated. As measured by the scratch-wound
healing assay, miR-191 mimic significantly enhanced cell monolayer
restoration in A549 at 24 h (Fig.
4A; P<0.01), while miR-191 inhibitor exhibited the opposite
effect (Fig. 4B; P<0.01). In
accordance with these findings, miR-191 mimic significantly
elevated the number of migratory cells through the Matrigel
basement membrane as observed by the Transwell invasion assay at 24
h (Fig. 5A; P<0.01), whereas
miR-191 inhibitor exhibited the opposite effect (Fig. 5B; P<0.01). Together, these data
indicated that miR-191 promotes the ability of A549 to migrate and
invade in vitro.
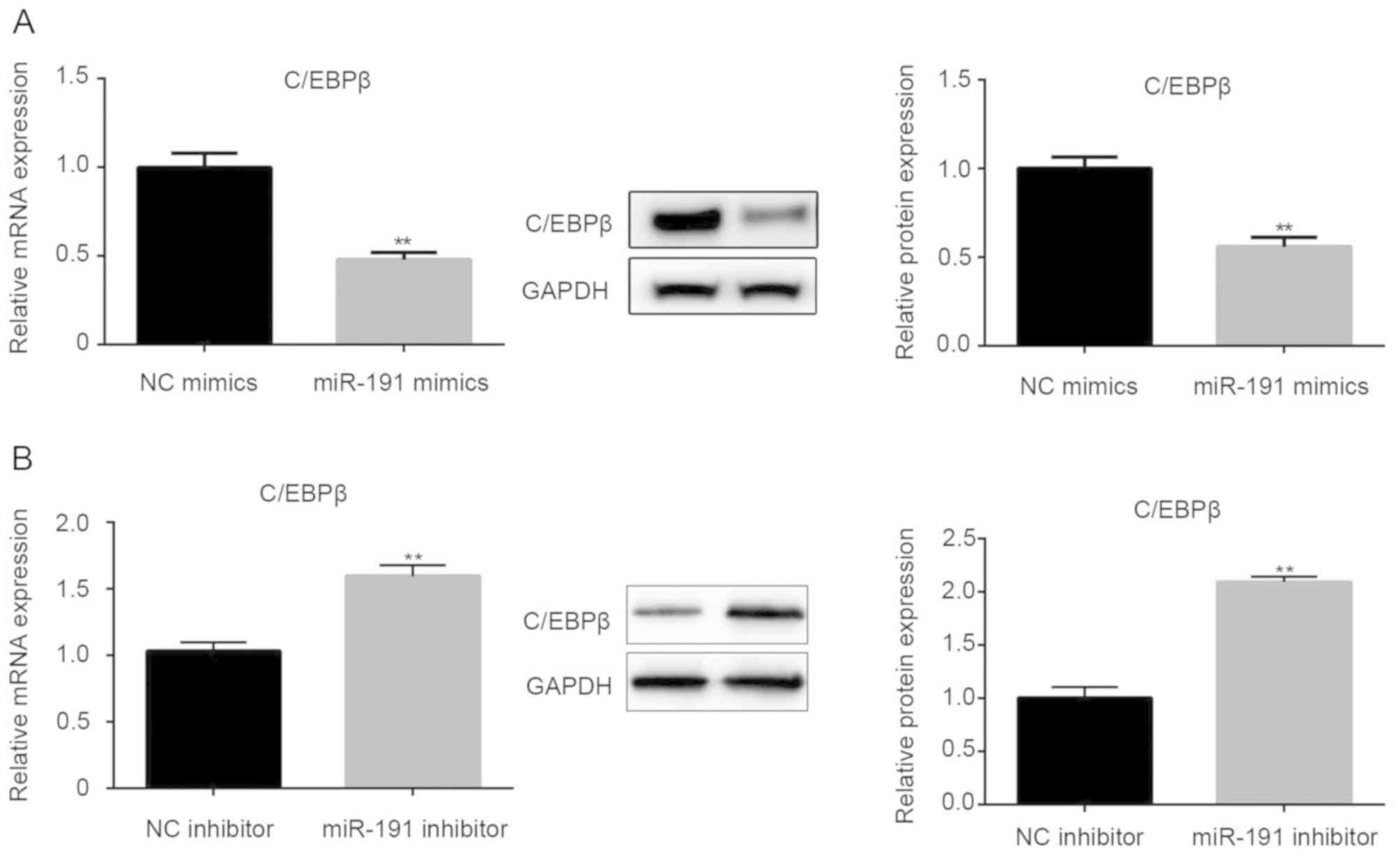
miR-191 inhibits the expression of
C/EBPβ
Based on bioinformatics analyses, C/EBPβ was
identified as a candidate target gene of miR-191. To explore the
regulation of C/EBPβ expression mediated by miR-191, RT-qPCR and
western blot assays were employed to detect the mRNA and protein
expression levels of C/EBPβ in miR-191-transfected A549 cells.
RT-qPCR analysis demonstrated that miR-191 significantly inhibited
mRNA and protein expression of C/EBPβ (Fig. 6A; P<0.01). Conversely, miR-191
inhibitor exhibited the opposite effects (Fig. 6B; P<0.01). These data suggested an
inverse association between miR-191 and C/EBPβ expression
levels.
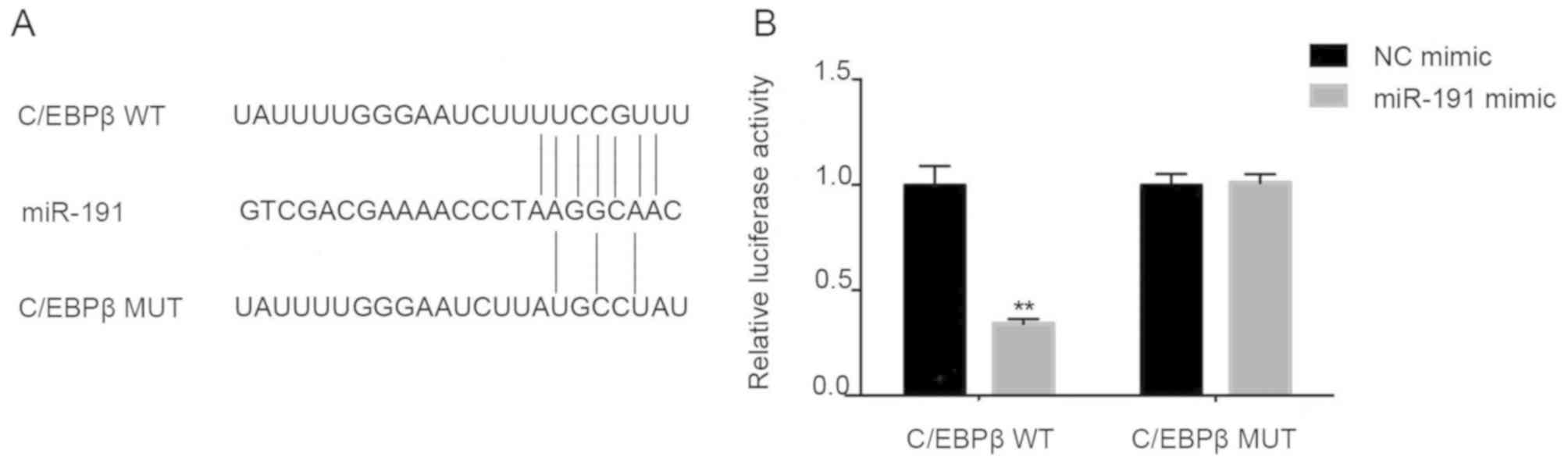
C/EBPβ is the direct target of
miR-191
Based on computational prediction, C/EBPβ has been
identified to be a direct target of miR-191 (http://www.targetscan.org and http://pictar.mdc-berlin.de/). Sequence analysis
revealed that C/EBPβ contains a putative binding site of miR-191
located in the 3′UTR (Fig. 7A). To
confirm the direct interaction between miR-191 and C/EBPβ, two
luciferase report plasmids were constructed, C/EBPβ 3′UTR wt and
C/EBPβ 3′UTR mut, and a dual luciferase reporter assay was
performed. Transfection with miR-191 mimic suppressed the
luciferase activity of C/EBPβ 3′UTR wt (Fig. 7B; P<0.01). Mutation of four
nucleotides in the predicted miR-191 binding site abolished the
inhibitory effects of miR-191 on luciferase activity. These results
provide evidence that miR-191 may have directly suppressed C/EBPβ
translation by specifically targeting the 3′UTR of C/EBPβ mRNA.
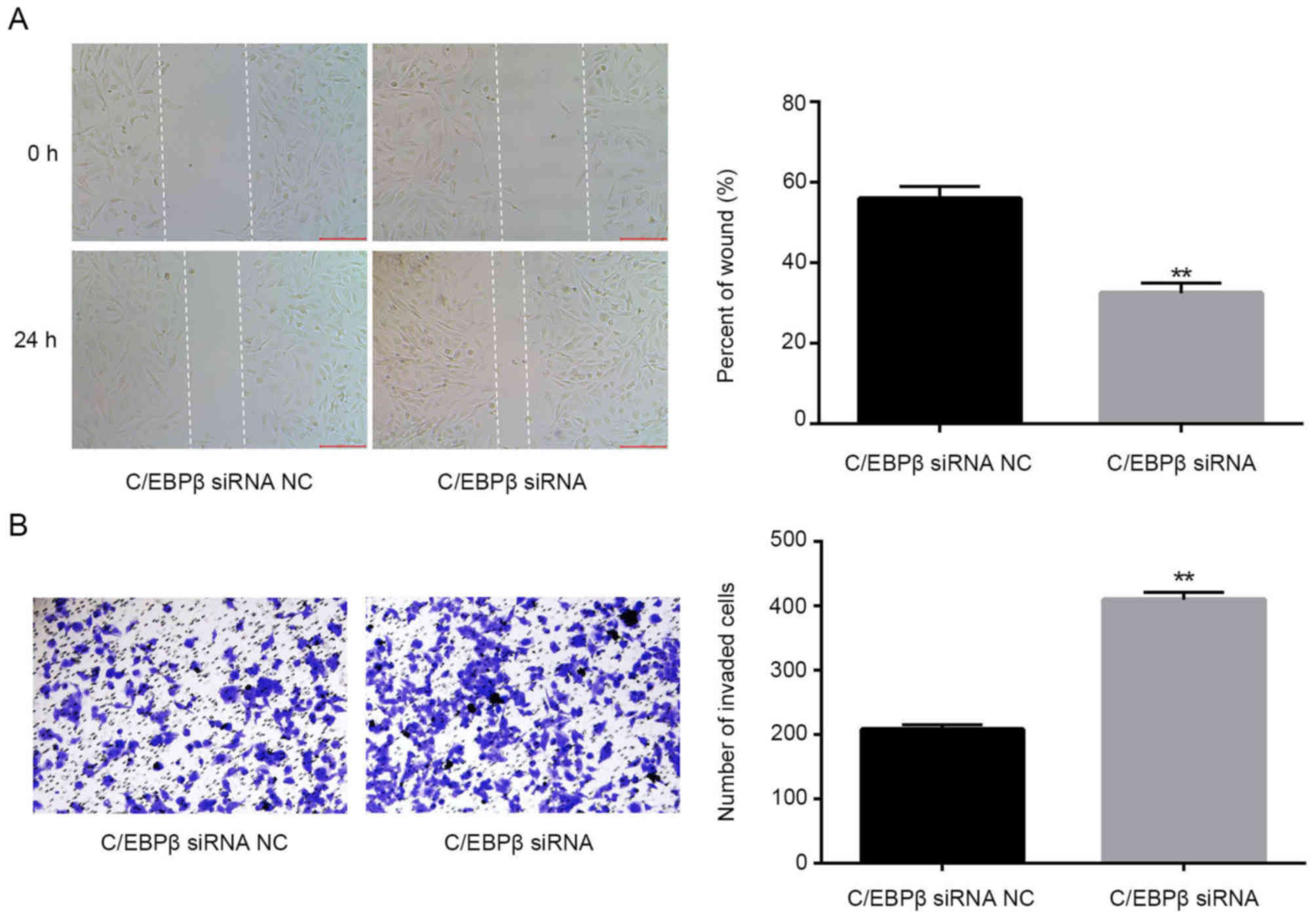
Knockdown of C/EBPβ mimics the effect
of miR-191 mimics
Finally, to determine the roles of C/EBPβ in lung
cancer, A549 cells were transfected with C/EBPβ siRNA, and the
migration and invasion of cells were examined. It was observed that
C/EBPβ siRNA could mimic the effects of miR-191 by promoting the
migration and invasion ability of A549 cells (Fig. 8A and B; P<0.01).
Discussion
Aberrant expression of miRNAs is linked with
multiple human cancers, indicating that miRNAs may be potential
therapeutic targets or biomarkers in human cancers as they can
modulate gene expression and a variety of cellular pathways
(27). Dysregulation of miRNAs can
disrupt tightly regulated RNA networks and thereby suppress or
enhance the progression of cancer (28). In this regard, the identification of
dysregulated miRNAs and their target genes may improve current
knowledge of the molecular mechanisms underlying tumorigenesis.
miR-191 has been reported to be upregulated in
multiple human cancers and may promote tumor growth and metastasis
(10,11,13). In
the present study, a significant upregulation of miR-191 was
observed in NSCLC tumor samples compared with in adjacent normal
tissue. Furthermore, it was observed that high expression levels of
miR-191 were associated with clinical stage and metastasis in
patients with NSCLC. Consistent with previous publications, these
findings strongly support the potential role of miR-191 as an
oncogenic gene in NSCLC. To clarify its role in NSCLC pathogenesis,
in vitro functional studies were performed in the NSCLC cell
line, A549. These results demonstrated that exogenous miR-191 may
promote the migration and invasion of A549 cells; however, the
underlying molecular mechanism of miR-191 in NSCLC remains
unclear.
Bioinformatics analysis identified C/EBPβ as a
direct target of miR-191. C/EBPβ may be involved in the regulation
of normal and cancer cell proliferation. For example, enhanced
expression of the C/EBPβ isoform liver-enriched inhibitory protein
was observed in advanced cases of breast and ovarian cancers, and
CRC (19,20,24).
Previously, an overall increase of in C/EBPβ expression was noted
in squamous cell carcinoma (29).
However, controversial findings have been reported in which the
aberrant expression of C/EBPβ promoted cell death in other cancers,
such as melanoma (30). The present
data support certain previous findings (30,31); it
was observed that the transfection with miR-191 mimic induced a
significant decrease in the expression of C/EBPβ, and transfection
of miR-191 inhibitor exhibited the opposite effects. Finally, the
dual luciferase assay confirmed the direct regulation of C/EBPβ
mediated by miR-191, and C/EBPβ siRNA can mimic the effects of
miR-191.
In conclusion, the present findings demonstrated a
positive association between miR-191 and NSCLC development. miR-191
may promote the migration and invasion of A549 cells in
vitro partially due to the regulation of its direct target
gene, C/EBPβ. The underlying mechanisms of C/EBPβ promoting the
invasion of A549 remain to be elucidated.
Acknowledgements
The authors would like to thank Anqiu People's
Hospital for their supervision of the current study (Anqiu,
China).
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL, JW and JS performed the experiments; FL, YW and
FY collected materials and interpreted the data; CL deigned and
approved the current study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and all study protocols were approved by the local ethics
committee of Anqiu People's Hospital (Anqiu, China).
Patient consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cui LH, Xu HR, Yang W and Yu LJ: lncRNA
PCAT6 promotes non-small cell lung cancer cell proliferation,
migration and invasion through regulating miR-330-5p. Onco Targets
Ther. 11:7715–7724. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li J and Wei L: Increased expression of
LINC01510 predicts poor prognosis and promotes malignant
progression in human non-small cell lung cancer. Biomed
Pharmacother. 109:519–529. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang B, Lv K, Chen W, Zhao J, Luo J, Wu J,
Li Z, Qin H, Wong TS, Yang W, et al: miR-375 and miR-205 regulate
the invasion and migration of laryngeal squamous cell carcinoma
synergistically via AKT-mediated EMT. Biomed Res Int.
2016:96527892016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagpal N, Ahmad HM, Molparia B and
Kulshreshtha R: MicroRNA-191, an estrogen-responsive microRNA,
functions as an oncogenic regulator in human breast cancer.
Carcinogenesis. 34:1889–1899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi X, Su S, Long J, Mei B and Chen Y:
MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and
promotes cell growth in human gastric carcinoma cell line MGC803.
Acta Biochim Biophys Sin (Shanghai). 43:849–856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Lu
J and Gao FH: MicroRNA-191 promotes pancreatic cancer progression
by targeting USP10. Tumour Biol. 35:12157–12163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Elyakim E, Sitbon E, Faerman A, Tabak S,
Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, et
al: hsa-miR-191 is a candidate oncogene target for hepatocellular
carcinoma therapy. Cancer Res. 70:8077–8087. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qin S, Zhu Y, Ai F, Li Y, Bai B, Yao W and
Dong L: MicroRNA-191 correlates with poor prognosis of colorectal
carcinoma and plays multiple roles by targeting tissue inhibitor of
metalloprotease 3. Neoplasma. 61:27–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J,
Shi BJ, Nie J, Ding XT, Li B, Zhou GW and Zhang ZY: MicroRNA-191
acts as a tumor promoter by modulating the TET1-p53 pathway in
intrahepatic cholangiocarcinoma. Hepatology. 66:136–151. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cruz-Munoz W and Khokha R: The role of
tissue inhibitors of metalloproteinases in tumorigenesis and
metastasis. Crit Rev Clin Lab Sci. 45:291–338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson GW, Johnson PF, Hennighausen L
and Sterneck E: The C/EBPbeta transcription factor regulates
epithelial cell proliferation and differentiation in the mammary
gland. Genes Dev. 12:1907–1916. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seagroves TN, Krnacik S, Raught B, Gay J,
Burgess-Beusse B, Darlington GJ and Rosen JM: C/EBPbeta, but not
C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar
proliferation, and functional differentiation in the mouse mammary
gland. Genes Dev. 12:1917–1928. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smink JJ, Bégay V, Schoenmaker T, Sterneck
E, de Vries TJ and Leutz A: Transcription factor C/EBPbeta isoform
ratio regulates osteoclastogenesis through MafB. EMBO J.
28:1769–1781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wethmar K, Bégay V, Smink JJ, Zaragoza K,
Wiesenthal V, Dörken B, Calkhoven CF and Leutz A:
C/EBPbetaDeltauORF mice-a genetic model for uORF-mediated
translational control in mammals. Genes Dev. 24:15–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang XF, Li KK, Gao L, Li SZ, Chen K,
Zhang JB, Wang D, Tu RF, Zhang JX, Tao KX, et al: miR-191 promotes
tumorigenesis of human colorectal cancer through targeting C/EBPβ.
Oncotarget. 6:4144–4158. 2015.PubMed/NCBI
|
|
20
|
Sundfeldt K, Ivarsson K, Carlsson M,
Enerbäck S, Janson PO, Brännström M and Hedin L: The expression of
CCAAT/enhancer binding protein (C/EBP) in the human ovary in vivo:
Specific increase in C/EBPbeta during epithelial tumour
progression. Br J Cancer. 79:1240–1248. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barakat DJ, Zhang J, Barberi T, Denmeade
SR, Friedman AD and Paz-Priel I: CCAAT/Enhancer binding protein β
controls androgen-deprivation-induced senescence in prostate cancer
cells. Oncogene. 34:5912–5922. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aguilar-Morante D, Morales-Garcia JA,
Santos A and Perez-Castillo A: CCAAT/enhancer binding protein β
induces motility and invasion of glioblastoma cells through
transcriptional regulation of the calcium binding protein S100A4.
Oncotarget. 6:4369–4384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gomis RR, Alarcón C, Nadal C, Van Poznak C
and Massagué J: C/EBPbeta at the core of the TGFbeta cytostatic
response and its evasion in metastatic breast cancer cells. Cancer
Cell. 10:203–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park BH, Kook S, Lee S, Jeong JH, Brufsky
A and Lee BC: An isoform of C/EBPβ, LIP, regulates expression of
the chemokine receptor CXCR4 and modulates breast cancer cell
migration. J Biol Chem. 288:28656–28667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang T, Cui M, Sun J, Ge C, Zhao F, Zhang
L, Tian H, Zhang L, Chen T, Jiang G, et al: Orosomucoid 2 inhibits
tumor metastasis and is upregulated by CCAAT/enhancer binding
protein β in hepatocellular carcinomas. Oncotarget. 6:16106–16119.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin SS, Peng CY, Liao YW, Chou MY, Hsieh
PL and Yu CC: miR-1246 targets CCNG2 to enhance cancer stemness and
chemoresistance in oral carcinomas. Cancers (Basel). 10(pii):
E2722018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen M, Wu L, Tu J, Zhao Z, Fan X, Mao J,
Weng Q, Wu X, Huang L, Xu M and Ji J: miR-590-5p suppresses
hepatocellular carcinoma chemoresistance by targeting YAP1
expression. EBioMedicine. 35:142–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anand S, Ebner J, Warren CB, Raam MS,
Piliang M, Billings SD and Maytin EV: C/EBP transcription factors
in human squamous cell carcinoma: Selective changes in expression
of isoforms correlate with the neoplastic state. PLoS One.
9:e1120732014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang X, Du T, Wang X, Zhang Y, Hu W, Du X,
Miao L and Han C: IDH1, a CHOP and C/EBPβ-responsive gene under ER
stress, sensitizes human melanoma cells to hypoxia-induced
apoptosis. Cancer Lett. 365:201–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan Y, Hanse EA, Stedman K, Benson JM,
Lowman XH, Subramanian S and Kelekar A: Transcription factor
C/EBP-β induces tumor-suppressor phosphatase PHLPP2 through
repression of the miR-17-92 cluster in differentiating AML cells.
Cell Death Differ. 23:1232–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|