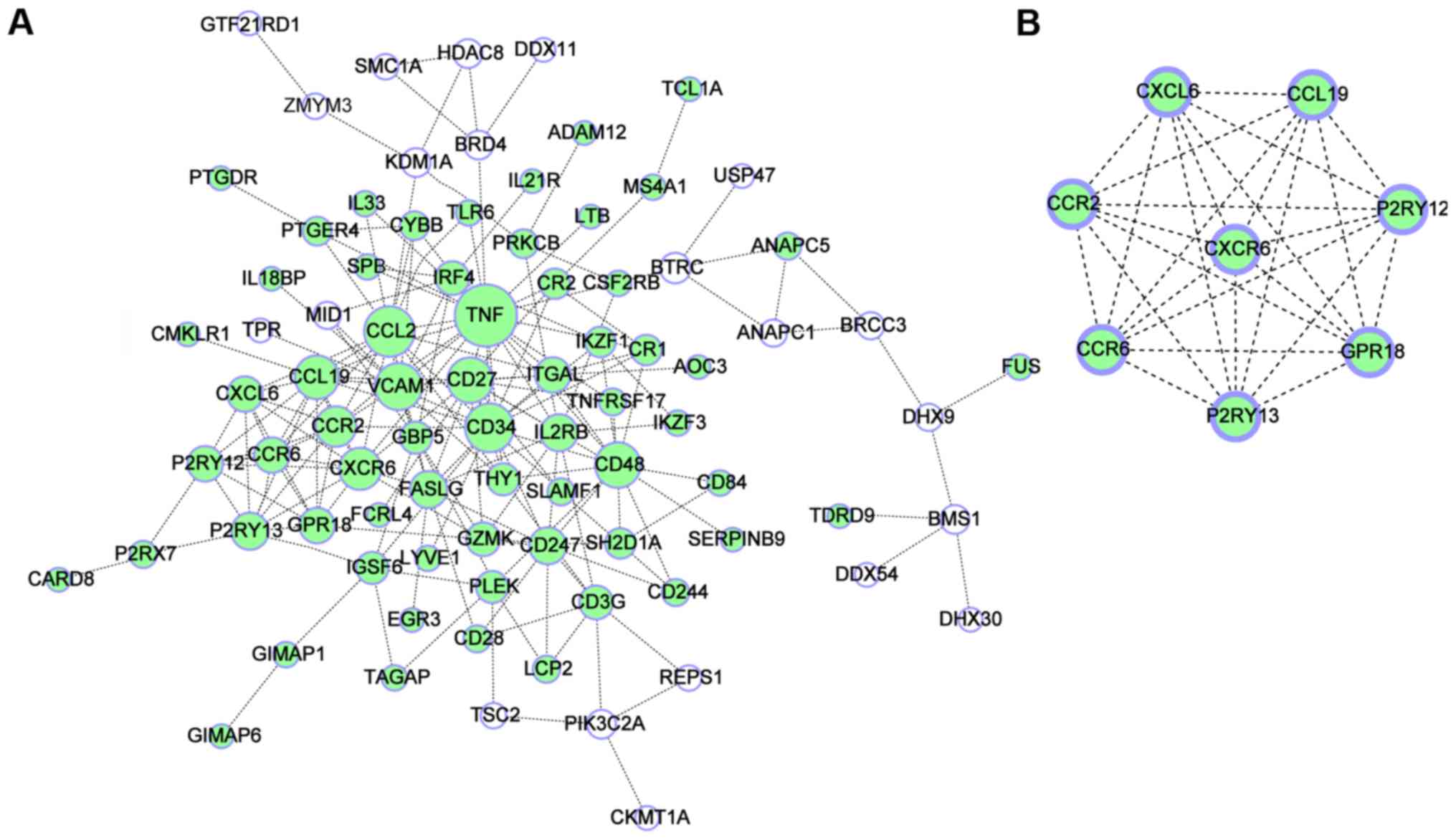
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chuang CH, Greenside PG, Rogers ZN, Brady
JJ, Yang D, Ma RK, Caswell DR, Chiou SH, Winters AF, Grüner BM, et
al: Molecular definition of a metastatic lung cancer state reveals
a targetable CD109-Janus kinase-Stat axis. Nat Med. 23:291–300.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tamura T, Kurishima K, Nakazawa K,
Kagohashi K, Ishikawa H and Hizawa N: Specific organ metastases and
survival in metastatic non-small-cell lung cancer. Mol Clin Oncol.
3:217–221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yano T, Yokoyama H, Inoue T, Asoh H,
Tayama K, Takai E and Ichinose Y: The first site of recurrence
after complete resection in non-small-cell carcinoma of the lung.
Comparison between pN0 disease and pN2 disease. J Thorac Cardiovasc
Surg. 108:680–683. 1994.PubMed/NCBI
|
|
5
|
Chen W, Zhang S and Zou X: Estimation and
projection of lung cancer incidence and mortality in China.
Zhongguo Fei Ai Za Zhi. 13:488–493. 2010.(In Chinese). PubMed/NCBI
|
|
6
|
Sandler A, Hirsh V, Reck M, von Pawel J,
Akerley W and Johnson DH: An evidence-based review of the incidence
of CNS bleeding with anti-VEGF therapy in non-small cell lung
cancer patients with brain metastases. Lung Cancer. 78:1–7. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Linardou H, Kalogeras KT, Kronenwett R,
Kouvatseas G, Wirtz RM, Zagouri F, Gogas H, Christodoulou C,
Koutras AK, Samantas E, et al: The prognostic and predictive value
of mRNA expression of vascular endothelial growth factor family
members in breast cancer: A study in primary tumors of high-risk
early breast cancer patients participating in a randomized Hellenic
Cooperative Oncology Group trial. Breast Cancer Res. 14:R1452012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen G, Liu XY, Wang Z and Liu FY:
Vascular endothelial growth factor C: The predicator of early
recurrence in patients with N2 non-small-cell lung cancer. Eur J
Cardiothorac Surg. 37:546–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen G, Wang Z, Liu XY and Liu FY:
High-level CXCR4 expression correlates with brain-specific
metastasis of non-small cell lung cancer. World J Surg. 35:56–61.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han CH and Brastianos PK: Genetic
characterization of brain metastases in the era of targeted
therapy. Front Oncol. 7:2302017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tarca AL, Romero R and Draghici S:
Analysis of microarray experiments of gene expression profiling. Am
J Obstet Gynecol. 195:373–388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wong SY and Hynes RO: Lymphatic or
hematogenous dissemination: How does a metastatic tumor cell
decide? Cell Cycle. 5:812–817. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Naxerova K, Reiter JG, Brachtel E, Lennerz
JK, van de Wetering M, Rowan A, Cai T, Clevers H, Swanton C, Nowak
MA, et al: Origins of lymphatic and distant metastases in human
colorectal cancer. Science. 357:55–60. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Law CW, Alhamdoosh M, Su S, Smyth GK and
Ritchie ME: RNA-seq analysis is easy as 1–2–3 with limma, Glimma
and edgeR. F1000Res. 5:14082016. View Article : Google Scholar
|
|
15
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Strogatz SH: Exploring complex networks.
Nature. 410:268–276. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mori T, Ikeda DD, Fukushima T, Takenoshita
S and Kochi H: NIRF constitutes a nodal point in the cell cycle
network and is a candidate tumor suppressor. Cell Cycle.
10:3284–3299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Preusser M, Winkler F, Valiente M,
Manegold C, Moyal E, Widhalm G, Tonn JC and Zielinski C: Recent
advances in the biology and treatment of brain metastases of
non-small cell lung cancer: Summary of a multidisciplinary
roundtable discussion. ESMO Open. 3:e0002622018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Whitsett TG, Inge LJ, Dhruv HD, Cheung PY,
Weiss GJ, Bremner RM, Winkles JA and Tran NL: Molecular
determinants of lung cancer metastasis to the central nervous
system. Transl Lung Cancer Res. 2:273–283. 2013.PubMed/NCBI
|
|
24
|
Duchnowska R, Pęksa R, Radecka B, Mandat
T, Trojanowski T, Jarosz B, Czartoryska-Arłukowicz B, Olszewski WP,
Och W, Kalinka-Warzocha E, Kozłowski W, et al: Immune response in
breast cancer brain metastases and their microenvironment: The role
of the PD-1/PD-L axis. Breast Cancer Res. 18:432016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berghoff AS, Venur VA, Preusser M and
Ahluwalia MS: Immune checkpoint inhibitors in brain metastases:
From biology to treatment. Am Soc Clin Oncol Educ Book.
35:e116–e122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zakaria R, Platt-Higgins A, Rathi N, Radon
M, Das S, Das K, Bhojak M, Brodbelt A, Chavredakis E, Jenkinson MD
and Rudland PS: T-cell densities in brain metastases are associated
with patient survival times and diffusion tensor MRI changes.
Cancer Res. 78:610–616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taggart D, Andreou T, Scott KJ, Williams
J, Rippaus N, Brownlie RJ, Ilett EJ, Salmond RJ, Melcher A and
Lorger M: Anti-PD-1/anti-CTLA-4 efficacy in melanoma brain
metastases depends on extracranial disease and augmentation of
CD8+ T cell trafficking. Proc Natl Acad Sci USA.
115:E1540–E1549. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woditschka S, Evans L, Duchnowska R, Reed
LT, Palmier D, Qian Y, Badve S, Sledge G Jr, Gril B, Aladjem MI, et
al: DNA double-strand break repair genes and oxidative damage in
brain metastasis of breast cancer. J Natl Cancer Inst.
106:diu1452014. View Article : Google Scholar
|
|
29
|
Fan Y, Zhu X, Xu Y, Lu X, Xu Y, Wang M, Xu
H, Ding J, Ye X, Fang L, et al: Cell-cycle and DNA-damage response
pathway is involved in leptomeningeal metastasis of non-small cell
lung cancer. Clin Cancer Res. 24:209–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pritchard CC, Mateo J, Walsh MF, De Sarkar
N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R,
et al: Inherited DNA-repair gene mutations in men with metastatic
prostate cancer. N Engl J Med. 375:443–453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Taher C, Frisk G, Fuentes S, Religa P,
Costa H, Assinger A, Vetvik KK, Bukholm IR, Yaiw KC, Smedby KE, et
al: High prevalence of human cytomegalovirus in brain metastases of
patients with primary breast and colorectal cancers. Transl Oncol.
7:732–740. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng ZH, Shi YX, Yuan M, Xiong D, Zheng
JH and Zhang ZY: Chemokines and their receptors in lung cancer
progression and metastasis. J Zhejiang Univ Sci B. 17:342–351.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Reckamp KL, Strieter RM and Figlin RA:
Chemokines as therapeutic targets in renal cell carcinoma. Expert
Rev Anticancer Ther. 8:887–893. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jassam SA, Maherally Z, Smith JR, Ashkg K,
Roncaroli F, Fillmore HL and Pilkington GJ: TNF-α enhancement of
CD62E mediates adhesion of non-small cell lung cancer cells to
brain endothelium via CD15 in lung-brain metastasis. Neuro Oncol.
18:679–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xing F, Sharma S, Liu Y, Mo YY, Wu K,
Zhang YY, Pochampally R, Martinez LA, Lo HW and Watabe K: miR-509
suppresses brain metastasis of breast cancer cells by modulating
RhoC and TNF-α. Oncogene. 34:4890–4900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Zhang S, Yao J, Lowery FJ, Zhang
Q, Huang WC, Li P, Li M, Wang X, Zhang C, et al:
Microenvironment-induced PTEN loss by exosomal microRNA primes
brain metastasis outgrowth. Nature. 527:100–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu X and Kang Y: Chemokine (C-C motif)
ligand 2 engages CCR2+ stromal cells of monocytic origin to promote
breast cancer metastasis to lung and bone. J Biol Chem.
284:29087–29096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng HW, Onder L, Cupovic J, Boesch M,
Novkovic M, Pikor N, Tarantino I, Rodriguez R, Schneider T, Jochum
W, et al: CCL19-producing fibroblastic stromal cells restrain lung
carcinoma growth by promoting local antitumor T-cell responses. J
Allergy Clin Immunol. 142:1257–1271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hillinger S, Yang SC, Zhu L, Huang M,
Duckett R, Atianzar K, Batra RK, Strieter RM, Dubinett SM and
Sharma S: EBV-induced molecule 1 ligand chemokine (ELC/CCL19)
promotes IFN-gamma-dependent antitumor responses in a lung cancer
model. J Immunol. 171:6457–6465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chung B, Esmaeili AA, Gopalakrishna-Pillai
S, Murad JP, Andersen ES, Kumar Reddy N, Srinivasan G, Armstrong B,
Chu C, Kim Y, et al: Human brain metastatic stroma attracts breast
cancer cells via chemokines CXCL16 and CXCL12. NPJ Breast Cancer.
3:62017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao Q, Zhao YJ, Wang XY, Qiu SJ, Shi YH,
Sun J, Yi Y, Sun JY, Shi GM, Ding ZB, et al: CXCR6 upregulation
contributes to a proinflammatory tumor microenvironment that drives
metastasis and poor patient outcomes in hepatocellular carcinoma.
Cancer Res. 72:3546–3556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Johnson LA, Vaidya SV, Goldfarb RH and
Mathew PA: 2B4 (CD244)-mediated activation of NK cells reduces
metastases of B16F10 melanoma in mice. Anticancer Res.
23:3651–3655. 2003.PubMed/NCBI
|
|
43
|
Pagès F, Berger A, Camus M, Sanchez-Cabo
F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte
D, et al: Effector memory T cells, early metastasis, and survival
in colorectal cancer. N Engl J Med. 353:2654–2666. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Simmons DL, Satterthwaite AB, Tenen DG and
Seed B: Molecular cloning of a cDNA encoding CD34, a sialomucin of
human hematopoietic stem cells. J Immunol. 148:267–271.
1992.PubMed/NCBI
|
|
45
|
Qiu H, Cao L, Wang D, Xu H and Liang Z:
High levels of circulating CD34+/VEGFR3+ lymphatic/vascular
endothelial progenitor cells is correlated with lymph node
metastasis in patients with epithelial ovarian cancer. J Obstet
Gynaecol Res. 39:1268–1275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lawson C, Ainsworth M, Yacoub M and Rose
M: Ligation of ICAM-1 on endothelial cells leads to expression of
VCAM-1 via a nuclear factor-kappaB-independent mechanism. J
Immunol. 162:2990–2996. 1999.PubMed/NCBI
|
|
47
|
Sato R, Nakano T, Hosonaga M, Sampetrean
O, Harigai R, Sasaki T, Koya I, Okano H, Kudoh J, Saya H and Arima
Y: RNA sequencing analysis reveals interactions between breast
cancer or melanoma cells and the tissue microenvironment during
brain metastasis. Biomed Res Int. 2017:80329102017. View Article : Google Scholar : PubMed/NCBI
|