Introduction
Esophageal cancer is a common malignancy of the
digestive tract, with the sixth-highest cancer associated mortality
rate worldwide (1). Esophageal
squamous cell carcinoma (ESCC) is particularly prevalent in
far-eastern countries including China (2). The 5-year survival rates of patients
with early esophageal cancer has improved drastically as a result
of multidisciplinary developments in treatment strategies; however,
the risk of mortality from advanced esophageal cancer remains
high.
Aberrant inactivation of tumor suppressor genes by
promoter methylation is frequently involved in tumorigenesis. In
particular, there has been accumulating evidence reveling a close
association between methylated tumor suppressor-encoding genes and
the development and progression of malignant tumors (3). Zinc finger proteins (ZFPs) are the
largest family of protein transcription factors in mammals. A
Krüppel-associated box zinc-finger protein, ZFP82/ZNF545/KIAA1948
is considered to be a novel tumor suppressive gene that is
frequently downregulated and methylated in multiple carcinomas
including in breast and gastric cancer (4). Furthermore, the biological functions of
ZNF545 are reported to be involved in cell proliferation and
apoptosis, and serve an important role in cancer development. It
has been demonstrated previously that ZNF545 suppresses tumor
progression by inhibiting nuclear factor-κB activation in ESCC cell
lines; however, it is frequently methylated in ESCC (4,5). In
addition, another previous study demonstrated that ZNF545 may be
involved in regulating p53 signaling in multiple myeloma (6).
Frequent point mutations in the tumor suppressor
gene p53 have been identified in both primary ESCCs and in ESCC
cell lines. Point mutations in this gene may occur at early stages
of ESCC and correlate with tumor progression, indicating that they
may serve an important role in ESCC. However, the role of ZNF545
remains unclear in p53-mutant ESCC cell lines.
Based on the role of ZNF545 in ESCC, the effect of
ZNF545 overexpression on p53 gene expression and its downstream
signaling pathways in the p53-mutant ESCC cell line, KYSE150, was
investigated. In addition, the clinical significance of ZNF545
expression in patients with ESCC was examined.
Materials and methods
Tissue specimens, cell line and
plasmid transfection
Fresh ESCC and adjacent normal tissues were obtained
from 64 patients (mean age, 63.1±7.5; 54 males; 10 females) with no
second primary malignant tumor that had undergone an esophagectomy
at the Department of Cardiothoracic Surgery, the Affiliated
Hospital of Southwest Medical University (Luzhou, China) from March
2015 to February 2016. All patients were newly diagnosed and had
not received treatment prior to the study. Written informed consent
was provided by all patients and the clinical and pathological data
were obtained. The current study was approved by the Institutional
Ethics Committees of the Affiliated Hospital of Southwest Medical
University, and followed the principles of the Declaration of
Helsinki. The esophageal cancer cell line, KYSE150, was anonymously
donated by a professor from Chongqing Medical University (Chonqing,
China) and was cultured in RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (PAA
Laboratories GmbH; GE healthcare) at 37°C in a humidified
atmosphere containing 5% CO2. The
pcDNA3.1(+)-Flag-ZNF545 plasmid was generated as previously
described (6). To establish stably
transfected cells with ectopic ZNF545 expression, the full-length
expression plasmid pcDNA3.1(+)-Flag-ZNF545 was transfected into
KYSE150 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at a final concentration of 4 µg.
The cells were then cultured and positively selected for resistance
in 350 µg/ml G418 (Sigma-Aldrich; Merck KGaA) for 14 days and
ZNF545 expression subsequently confirmed via semi-quantitative
RT-PCR and western blotting.
DNA and RNA extraction
Total RNA was isolated from KYSE150 cells and
esophageal tissues using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), whereas genomic DNA was extracted from KYSE150
cells and esophageal tissues using a QIAamp DNA Mini kit (Qiagen
GmbH) according to the manufacturer's protocol. DNA and RNA
integrity were assessed using gel electrophoresis, and the nucleic
acid samples were stored at −80°C until required.
5-Aza-2′-deoxycytidine (Aza) and
trichostatin A (TSA) treatment
KYSE150 cells were seeded into 100 mm dishes at a
density of 1×105 cells/ml. Cells were first treated with
10 mmol/l of the demethylating reagent, Aza (Sigma-Aldrich; Merck
KGaA), for 3 days at 37°C, before the addition of 100 nmol/l
histone deacetylase inhibitor, TSA (Sigma-Aldrich; Merck KGaA), for
a further 24 h at 37°C in a humidified atmosphere containing 5%
CO2, as described previously (5). RNA was subsequently collected from
cells as aforementioned for the analysis of ZNF545 mRNA
expression.
Semi-quantitative reverse
transcription (RT)-PCR
RNA was quantified using the NanoDrop ND2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). The A260/A280 nm ratio was calculated as between 1.6 and
1.9. Subsequently, cDNA was synthesized from 1 µg total RNA using
the reverse transcription (RT) system (Promega Corporation), and
was stored at −20°C until required. Semi-quantitative RT-PCR for
ZNF545 was performed using treated/untreated KYSE150 cells as
described previously (6). PCR
products were amplified with 32 cycles for ZNF545 and 23 cycles for
β-actin using the following temperature protocol: Initial
denaturation at 95°C for 10 min, denaturation at 95°C for 30 sec,
annealing at 55°C for 30 sec, elongation at 72°C for 30 sec and
final extension at 25°C for 10 min. Amplification was performed
using Go-Taq® polymerase (Promega Corporation) and
products were assayed on 2% agarose gels. Samples were subsequently
visualized and analyzed using the ChemiDoc MP system and Image Lab
3.0 software (each, Bio-Rad Laboratories, Inc.).
RT-quantitative PCR (RT-qPCR)
GoTaq qPCR Master Mix (Promega Corporation) was used
for RT-qPCR analysis, which was performed using a 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.),
following which melting-curve analysis was performed. Relative
target gene expression was calculated using the 2−ΔΔCq
method (7) with β-actin applied as
the internal reference. The primer sequences are listed in Table I.
 | Table I.List of primers used in the present
study. |
Table I.
List of primers used in the present
study.
| Primers | Sequences
(5′-3′) | Product size
(bp) |
|---|
| ZNF545-F |
GAGCCTTGGAAAGTTGTGAG | 245 |
| ZNF545-R |
GGCATTTTCACACTACTGAAG |
|
| p53-F |
TCAACAAGATGTTTTGCCAACTG | 118 |
| p53-R |
ATGTGCTGTGACTGCTTGTAGATG |
|
| β-actin-F |
CCTGTGGCATCCACGAAACT | 314 |
| β-actin-R |
GAAGCATTTGCGGTGGACGAT |
|
| ZNF545-m1-F |
TTTTTTTTAGGTTTTGTCGCGTC | 177 |
| ZNF545-m2-R |
CTACTAAAAAAACCGAACGCG |
|
| ZNF545-u1-F |
TTTTTTTTTAGGTTTTGTTGTGTT | 164 |
| ZNF545-u2-R |
CCAAACACACTCACAAAATACA |
|
Bisulfite modification of DNA and
methylation-specific PCR (MSP)
Bisulfite modification of DNA extracted from ESCC
tissue and MSP were performed as described previously (8). DNA treated with bisulfite was amplified
using MSP with methylation-specific (M1 and M2) or
non-methylation-specific (U1 and U2) primer pairs. The primer
sequences are listed in Table I. MSP
was performed for 40 cycles with the following thermocycling
conditions: Initial denaturation at 95°C for 10 min, denaturation
at 95°C for 30 sec, annealing at 58°C for 30 sec, elongation at
72°C for 30 sec and final extension at 72°C for 10 min. MSP was
performed using AmpliTaq-Gold DNA Polymerase (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with annealing temperatures of 60°C
for M1/M2 and 58°C for U1 and U2. The MSP products were identified
by performing gel electrophoresis with 2% agarose gels and a 100 bp
DNA ladder as a marker (MBI Fermentas; Thermo Fisher Scientific,
Inc.). The gels were visualized and analyzed as aforementioned.
Colony formation assay
KYSE150 cells stably transfected with
pcDNA3.1(+)-Flag-ZNF545 or empty vector were seeded into a six-well
plate at a density of 800 cells/well in culture media supplemented
with G418. Following 14–21 days, cells were fixed using 4%
paraformaldehyde for 30 min at room temperature and subsequently
stained using 0.1% crystal violet solution for 15 min at room
temperature. Colonies with >50 cells/colony were counted. The
assay was performed three times for triplicate samples.
Flow cytometry analysis of
apoptosis
To measure apoptotic cells, 2×105 cells
were harvested with 0.1% trypsin from 6-well plates at 48 h
following transfection with the ZNF545 plasmid and empty controls,
and centrifugation at 400 × g for 5 min at room temperature.
Apoptosis was measured by staining the cells with Annexin
V-fluorescein isothiocyanate (FITC) and propidium iodide (PI; BD
Biosciences) according to the manufacturer's protocol, and was
analyzed using a Navios flow cytometer with Kaluza 2.0 software
(Beckman Coulter, Inc.). All experiments were performed three times
for triplicate samples.
Western blot analysis
At 48 h following transfection, cells were lysed
using Mammalian Protein Extraction Reagent (Thermo Fisher
Scientific, Inc.) supplemented with a phosphatase inhibitor
cocktail (Sigma-Aldrich; Merck KGaA). Protein concentrations were
determined using bicinchoninic acid Protein Assay Reagent (Thermo
Fisher Scientific, Inc.). The protein samples were subsequently
denatured and 40 µg of each sample was separated by 10% SDS-PAGE
before transferal onto PVDF membranes (Bio-Rad Laboratories, Inc.)
(6). Following blocking with 5%
non-fat milk at room temperature for 2 h, and washing using TBS
supplemented with 0.1% Tween-20, membranes were incubated overnight
at 4°C with the following primary antibodies: ZNF545 (cat. no.
sc-102235; 1:1,000; Santa Cruz Biotechnology, Inc.), Akt (cat. no.
4691; 1:1,000; Cell Signaling Technology, Inc.), phosphorylated
(p)-Akt (cat. no. 4060; 1:2,000; Cell Signaling Technology, Inc.),
p53 (cat. no. sc-126; 1:5,000; Santa Cruz Biotechnology, Inc.),
BCL2 associated X, apoptosis regulator (Bax; cat. no. 5023;
1:1,000; Cell Signaling Technology, Inc.), p21 (cat. no. 2946;
1:2,000; Cell Signaling Technology, Inc.), cleaved-caspase 3 (cat.
no. 9665; 1:1,000; Cell Signaling Technology, Inc.),
cleaved-caspase 7 (cat. no. 9491; 1:1,000; Cell Signaling
Technology, Inc.) and cleaved-caspase 9 (cat. no. 7237; 1:1,000;
Cell Signaling Technology, Inc.), with β-actin (cat. no.
LK-ab008-100; 1:200; Hangzhou Lianke Biology Technology Co., Ltd.)
used as the internal loading control. Horseradish peroxidase
(HRP)-conjugated anti-rabbit IgG (cat. no. 7074; 1:2,000; Cell
Signaling Technology, Inc.) and HRP-conjugated anti-mouse IgG (cat.
no. 7076; 1:2,000; Cell Signaling Technology, Inc.) secondary
antibodies were added and incubated in a shaker at room temperature
for 60 min. Protein blots were visualized using an enhanced
chemiluminescence kit (Amersham Pharmacia Biotech Ltd.; GE
Healthcare) and imaged and analyzed using the LAS-4000 Imaging
System (Medical Systems; Fujifilm).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 13.0; SPSS, Inc.). All data were presented as the
mean ± standard deviation and analyzed using Student's t-test,
Wilcoxon signed-rank tests and Pearson's chi-squared tests as
appropriate. All experiments were repeated in triplicate and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Promoter methylation downregulates
ZNF545 expression in ESCC tissues
The expression of ZNF545 mRNA in tumor and paired
adjacent normal tissues from 15 patients diagnosed with ESCC was
examined using RT-qPCR. ZNF545 mRNA was significantly downregulated
in ESCC tumors when compared with adjacent normal tissues (P=0.022;
Fig. 1A). ZNF545 expression was also
restored following the pharmacologic treatment of Aza and TSA in
KYSE150 (Fig. 1B). The promoter
methylation status of ZNF545 in tumor tissues (n=64) and adjacent
normal tissues (n=64) was also evaluated using an MSP assay. ZNF545
promoter methylation was observed in 76.6% (49/64) of primary
tumors, compared with only 28.1% (28/64) of adjacent normal tissues
(P<0.001; Fig. 1C and Table II). Lastly, the association between
ZNF545 methylation and clinicopathological features in ESCC
patients was investigated, but no significant association was
observed for age, sex, tumor location, staging, pathologic grade or
lymph node metastasis (P>0.05 for all features; Table III).
 | Table II.Promoter methylation status of ZNF545
in esophageal cancer. |
Table II.
Promoter methylation status of ZNF545
in esophageal cancer.
|
|
| ZNF545 promoter |
|
|---|
|
|
|
|
|
|---|
| Tissues | Samples (n) | Methylated | Unmethylated | Frequency of
methylation (%) |
|---|
| Esophageal
tumors | 64 | 49 | 15 | 76.6a |
| Adjacent esophageal
tissues | 64 | 18 | 46 | 28.1 |
 | Table III.Association between ZNF545 methylation
and the clinicopathological features of patients with esophageal
cancer. |
Table III.
Association between ZNF545 methylation
and the clinicopathological features of patients with esophageal
cancer.
|
|
| ZNF545 methylation
status |
|
|---|
|
|
|
|
|
|---|
| Clinicopathologic
feature | Number (n=64) | Methylated | Unmethylated | P-value |
|---|
| Age |
|
|
| 0.447 |
| ≤60 | 23 | 19 | 4 |
|
|
>60 | 38 | 27 | 11 |
|
|
Unknown | 3 | 3 | 0 |
|
| Sex |
|
|
| 0.731 |
|
Male | 51 | 39 | 12 |
|
|
Female | 10 | 7 | 3 |
|
|
Unknown | 3 | 3 | 0 |
|
| Tumor location |
|
|
| 0.596 |
|
Upper | 13 | 9 | 4 |
|
|
Median | 35 | 26 | 9 |
|
|
Lower | 24 | 20 | 4 |
|
|
Unknown | 4 | 4 | 0 |
|
| Tumor stage |
|
|
| 0.613 |
|
I–II | 27 | 22 | 5 |
|
|
III–IV | 19 | 13 | 6 |
|
|
Unknown | 18 | 14 | 4 |
|
| Pathologic
grade |
|
|
| 0.158 |
| G1 | 25 | 22 | 3 |
|
| G2 | 37 | 24 | 13 |
|
| G3 | 11 | 7 | 4 |
|
|
Unknown | 6 | 4 | 2 |
|
| Lymph node
metastasis |
|
|
| 0.368 |
|
Positive | 13 | 8 | 5 |
|
|
Negative | 48 | 38 | 10 |
|
|
Unknown | 3 | 3 | 0 |
|
ZNF545 transfection inhibits cell
clonogenicity and induces apoptosis in ESCC cells
ZNF545 function in ESCC was evaluated by ectopically
overexpressing ZNF545 in the ESCC cell line, KYSE150 (Fig. 2A). The effect of ZNF545 expression on
KYSE150 cell proliferation was subsequently determined using a
colony formation assay. The total number of colonies was
significantly decreased in cells stably transfected with the ZNF545
vector (P<0.001) compared with those transfected with the
control vector (Fig. 2B). The
effects of ectopic ZNF545 expression on apoptosis was then examined
in KYSE150 cells using annexin V-FITC/PI double-staining. A
significant increase in the number of apoptotic cells was observed
in the ZNF545 overexpression group when compared with controls
(P<0.001; Fig. 2C). In support of
this, a marked increase in cleaved caspases 3, 7 and 9 protein
expression levels were also observed in KYSE150 cells ectopically
expressing ZNF545 when compared with those transfected with the
control vector (Fig. 2D).
ZNF545 suppresses tumors via the
transcriptional activation of p53 or a p53-independent signaling
pathway in p53-mutant KYSE150 cells
In a previous study, ZNF545 was identified as a
novel tumor suppressor that inhibited cell proliferation and colony
formation, and induced apoptosis via TP53 reactivation in multiple
myeloma (6). The association between
ZNF545 and the p53 signaling pathway was examined as a consequence
of the potential tumor suppressive function of ZNF545 revealed in
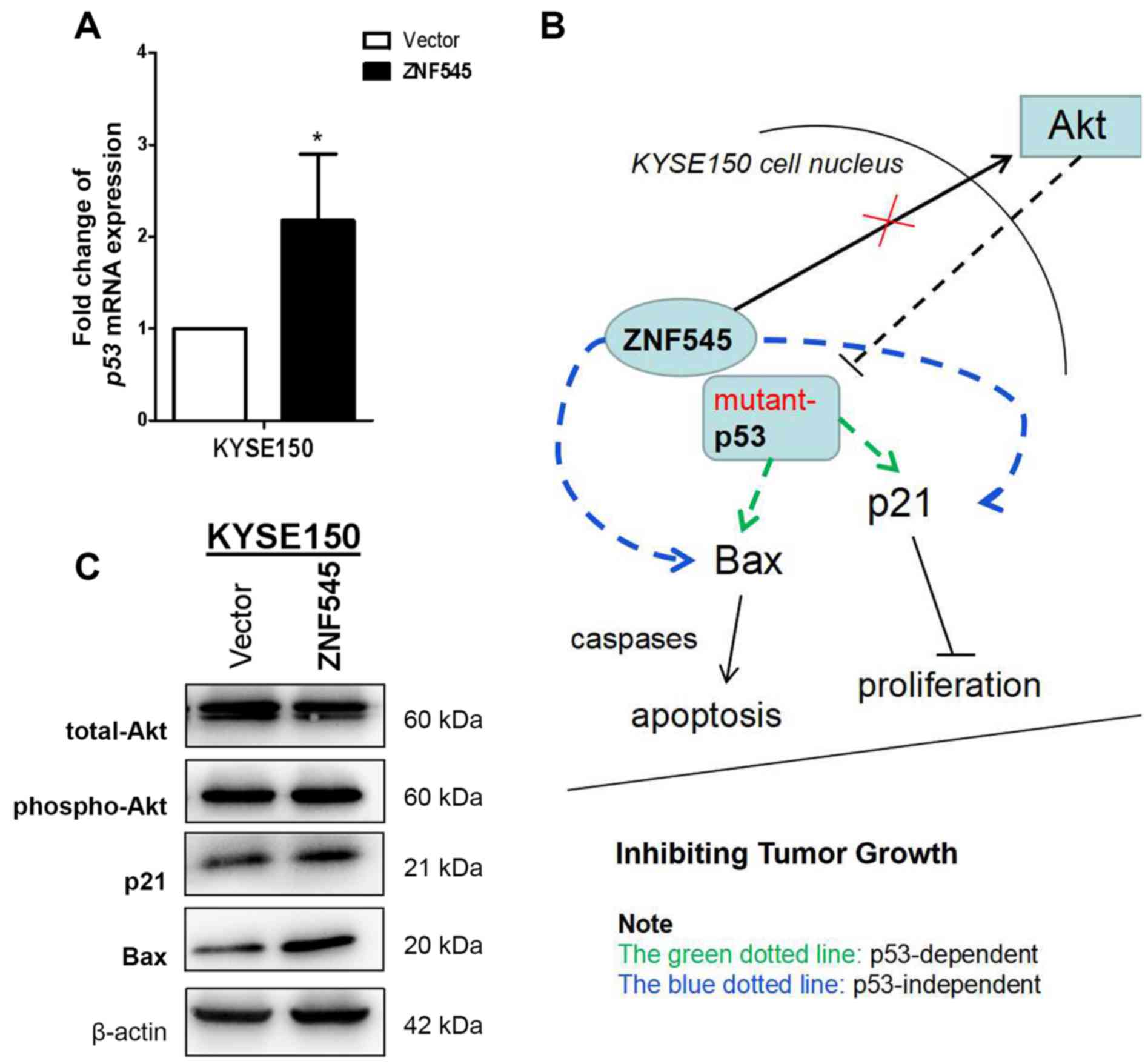
ESCC. RT-qPCR analysis revealed that ectopic ZNF545 expression in
KYSE150 cells upregulated p53 mRNA expression when compared with
those transfected with the control vector (P<0.05; Fig. 3A), suggesting that ZNF545 may
activate p53 transcription. The protein expression levels of two
downstream effectors of the p53 pathway, Bax and p21, were
increased in KYSE150 p53-mutant cells transfected with ZNF545;
whereas the expression of Akt was not affected by ZNF545
transfection (Fig. 3C). Although
ZNF545 may exert its tumor suppressive effects in ESCC by
activating p53 transcription through a p53-dependent signaling
pathway, intracellular signaling remains unclear. Furthermore,
upregulating p21 and Bax expression via p53-independent signaling
pathways in tumor cells with mutant p53 is another possibility
(Fig. 3B).
Discussion
Previous studies have identified ZNF545 as a
functional tumor suppressor gene in a number of solid tumors,
including ESCC, and its expression is frequently silenced by
promoter methylation (4,5,9,10). However, the association between p53
and ZNF545, and its downstream functions in p53-mutant ESCC remain
unclear. The present study provides novel evidence for an
association between ZNF545 and mutant p53 in ESCC. To assess ZNF545
methylation as a potential epigenetic biomarker, its relevance in
relation to the clinicopathological features of ESCC was also
evaluated, as aberrant DNA methylation has been reported to mediate
tumor suppressor gene silencing and consequent tumorigenesis
(11). As demonstrated by MSP
analysis in the present study, ZNF545 methylation was detected in
76.6% of ESCC tissues when compared with 28.1% of adjacent normal
tissues. However, no significant association between the
clinicopathological features of patients with ESCC and the
methylation status of ZNF545 was observed. In general, bisulfite
sequencing PCR (BSP) is applied to discover methylation sites on
genes, and MSP primers are subsequently designed according to the
methylation sites to determine the optimal conditions and methods
to detect methylation. BSP can yield accurate results due to
subsequent sequencing analysis; however, a high number of clones
are required for this, making the procedure more difficult to
operate in large quantities. MSP can be used to screen a large
number of specimens for the detection of methylation status.
Therefore, the MSP was selected to evaluate ZNF545 methylation in
clinical samples in the present study. A previous study
demonstrated that ZNF545 methylation was associated with survival
in patients with ESCC (5), and poor
survival in gastric and hepatic cancer (12,13). The
present study also revealed that ectopic ZNF545 expression
inhibited cell growth by significantly suppressing colony formation
and inducing apoptosis in ESCC cells.
The biological function of p53 in tumor suppression
is well established (14). p53
activates downstream mediators of cell proliferation inhibition
(p21 and growth arrest and DNA damage inducible-α) and
pro-apoptotic proteins (Bax and PUMA) resulting in cell cycle
arrest and apoptosis (15,16). In addition, the zinc finger protein,
JAZ, has been reported to serve a role in mediating tumor growth by
regulating p53 activity (17).
Transcriptional activation of TP53 by ZNF545 has also been reported
in breast cancer (10). Although
activation of the MDM2 ubiquitin-ligase by Akt and subsequent p53
degradation have been implicated in tumorigenesis (18,19), no
changes in Akt expression following ectopic ZNF545 expression was
observed in ESCC cells in the present study, which is consistent
with previous observations in multiple myeloma (6). However, p53 mutations are more commonly
observed in malignancies involving organs compared with their
hematological counterparts (20).
Strategies targeting mutant p53, and altered signaling pathways
exhibited in p53-mutant cells have been evaluated with the aim of
uncovering novel treatments for malignancies associated with mutant
p53 (20).
The results of the present study suggest that ZNF545
may transcriptionally activate p53 and upregulate the protein
expression of its potential downstream effectors, p21 and Bax, in
the p53-mutant KYSE150 ESCC cell line. As the functional status of
p53 in KYSE150 cells is unknown, these observations indicate that
mutant p53 proteins in ESCC may retain the ability to activate p21,
but also suggests that ZNF545-mediated tumor suppression may be
independent of p53 (21), thus
providing insights into a potentially novel tumorigenic mechanism
in ESCC. Indeed, p21 can be regulated by p53-dependent and
p53-independent pathways (22).
However, the mechanism by which ZNF545 may upregulate p21 and other
effectors in the absence of p53 warrants further evaluation. One
hypothesis is that the function of mutant p53 could be restored by
specific DNA binding or transcriptional transactivation via a
p53-dependent pathway (23).
Alternatively, it is possible that p21 and Bax may be upregulated
by ZNF545 through an as yet undiscovered p53-independent signaling
pathway, such as the nuclear factor-κB signaling pathway (24). A further in-depth investigation of
the role of ZNF545 in mutant-p53 tumors, and the potential
crosstalk with other signaling pathways, will need to be performed
to support the results of the present study.
In conclusion, the anti-tumor effects of ZNF545 in a
p53-mutant ESCC cell line may be mediated by the transcriptional
activation of p53 or by the induction of p21 and Bax by an as yet
unknown p53-independent signaling pathway. In addition, activation
of these potential signaling pathways during tumorigenesis may be
inhibited by ZNF545 methylation. Tumor-specific methylation of
ZNF545 may be a potential epigenetic biomarker for the early
diagnosis of ESCC. However, data in the present study is limited by
the lack of additional ESCC cell lines harboring other mutant-p53
genotypes to confirm these findings. In addition, no other ESCC
cell line with upregulated expression was available to perform a
ZNF545 knockdown assay.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research
Foundation of Southwest Medical University for Youth (grant no.
0903-00030685), the Funded Project of Affiliated Hospital of
Southwest Medical University for Doctors (grant no. 17135), the
Health and Family Planning Commission of Sichuan Province (grant
no. 17PJ557) and the National Natural Science Foundation of China
(grant no. 81201682).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF designed this study, contributed to the
experiments, and drafted the manuscript. YW contributed to the
experiments and interpreted the results. SF performed the data
analysis. DL and SL contributed to the acquisition of clinical
samples. All authors contributed to the revisions of this
manuscript and approved the final draft.
Ethics approval and consent to
participate
Ethics approval was received from the Affiliated
Hospital of Southwest Medical University. All participants provided
written consent during their enrollment.
Patient consent for publication
All patients provided written consent during
enrollment for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Murphy G, McCormack V, Abedi-Ardekani B,
Arnold M, Camargo MC, Dar NA, Dawsey SM, Etemadi A, Fitzgerald RC,
Fleischer DE, et al: International cancer seminars: A focus on
esophageal squamous cell carcinoma. Ann Oncol. 28:2086–2093. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holubekova V, Mendelova A, Jasek K,
Mersakova S, Zubor P and Lasabova Z: Epigenetic regulation by DNA
methylation and miRNA molecules in cancer. Future Oncol.
13:2217–2222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng Y, Liang P, Geng H, Wang Z, Li L,
Cheng SH, Ying J, Su X, Ng KM, Ng MH, et al: A novel 19q13
nucleolar zinc finger protein suppresses tumor cell growth through
inhibiting ribosome biogenesis and inducing apoptosis but is
frequently silenced in multiple carcinomas. Mol Cancer Res.
10:925–936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye L, Xiang T, Fan Y, Zhang D, Li L, Zhang
C, He X, Xiang Q, Tao Q and Ren G: The 19q13 KRAB Zinc-finger
protein ZFP82 suppresses the growth and invasion of esophageal
carcinoma cells through inhibiting NF-κB transcription and inducing
apoptosis. Epigenomics. 11:65–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fan Y, Zhan Q, Xu H, Li L, Li C, Xiao Q,
Xiang S, Hui T, Xiang T and Ren G: Epigenetic identification of
ZNF545 as a functional tumor suppressor in multiple myeloma via
activation of p53 signaling pathway. Biochem Biophys Res Commun.
474:660–666. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao Q, Huang H, Geiman TM, Lim CY, Fu L,
Qiu GH and Robertson KD: Defective de novo methylation of viral and
cellular DNA sequences in ICF syndrome cells. Hum Mol Genet.
11:2091–2102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang
H, Kang W, Li X, Tao Q, Sung JJ and Yu J: Zinc-finger protein 545
is a novel tumour suppressor that acts by inhibiting ribosomal RNA
transcription in gastric cancer. Gut. 62:833–841. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao Y, Xiang T, Luo X, Li C, Li Q, Peng
W, Li L, Li S, Wang Z, Tang L, et al: Zinc-finger protein 545
inhibits cell proliferation as a tumor suppressor through inducing
apoptosis and is disrupted by promoter methylation in breast
cancer. PLoS One. 9:e1109902014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng J, Liang H, Ying G, Dong Q, Zhang R,
Yu J, Fan D and Hao X: Poor survival is associated with the
methylated degree of zinc-finger protein 545 (ZNF545) DNA promoter
in gastric cancer. Oncotarget. 6:4482–4495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Li X, Tao Q, Yu XL, Cheng ZG, Han
ZY, Guo M and Liang P: Hypermethylation of ZNF545 is associated
with poor prognosis in patients with early-stage hepatocellular
carcinoma after thermal ablation. Gut. 64:1836–1837. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaiser AM and Attardi LD: Deconstructing
networks of p53-mediated tumor suppression in vivo. Cell Death
Differ. 25:93–103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shu KX, Li B and Wu LX: The p53 network:
P53 and its downstream genes. Colloids Surf B Biointerfaces.
55:10–18. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ozaki T, Nakagawara A and Nagase H: RUNX
Family participates in the regulation of p53-dependent DNA damage
response. Int J Genomics. 2013:2713472013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang M, Wu S, Su X and May WS: JAZ
mediates G1 cell-cycle arrest and apoptosis by positively
regulating p53 transcriptional activity. Blood. 108:4136–4145.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gottlieb TM, Leal JF, Seger R, Taya Y and
Oren M: Cross-talk between Akt, p53 and Mdm2: Possible implications
for the regulation of apoptosis. Oncogene. 21:1299–1303. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janicke RU, Sohn D and Schulze-Osthoff K:
The dark side of a tumor suppressor: Anti-apoptotic p53. Cell Death
Differ. 15:959–976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao D, Tahaney WM, Mazumdar A, Savage MI
and Brown PH: Molecularly targeted therapies for p53-mutant
cancers. Cell Mol Life Sci. 74:4171–4187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Andries V, Vandepoele K, Staes K, Berx G,
Bogaert P, Van Isterdael G, Ginneberge D, Parthoens E,
Vandenbussche J, Gevaert K and van Roy F: NBPF1, a tumor suppressor
candidate in neuroblastoma, exerts growth inhibitory effects by
inducing a G1 cell cycle arrest. BMC Cancer. 15:3912015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mirzayans R, Andrais B, Kumar P and Murray
D: Significance of Wild-type p53 signaling in suppressing apoptosis
in response to chemical genotoxic agents: Impact on chemotherapy
outcome. Int J Mol Sci. 18(pii): E9282017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Selivanova G and Wiman KG: Reactivation of
mutant p53: Molecular mechanisms and therapeutic potential.
Oncogene. 26:2243–2254. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Basile JR, Eichten A, Zacny V and Münger
K: NF-kappaB-mediated induction of p21(Cip1/Waf1) by tumor necrosis
factor alpha induces growth arrest and cytoprotection in normal
human keratinocytes. Mol Cancer Res. 1:262–270. 2003.PubMed/NCBI
|