Introduction
Breast cancer is the second most common malignant
tumor which affects 1.7 million individuals annually worldwide,
accounting for ~12% of all patients with cancer (1). Despite its favorable prognosis, breast
cancer is the fifth leading cause of cancer-associated mortality
and is responsible for >500,000 deaths every year worldwide
(2). Breast cancer is divided into
molecular subtypes based on the expression of three tumor markers,
which include the progesterone receptor (PR), estrogen receptor
(ER) and human epidermal growth factor 2-neu (HER2) (3,4). These
are frequently detected to guide clinical treatment and to verify
bioinformatics (3,4). As a major molecular subtype of breast
cancer, triple negative breast cancer is characterized by the
absence of ER and PR, the lack of HER2 overexpression and a poor
prognosis (5). Furthermore, the
treatment of triple negative breast cancer is limited due to the
heterogeneous nature of this disease, which therefore results in an
unclear pathogenesis (6).
Glucose metabolism serves a pivotal role in the
growth and development of normal and cancerous cells that exhibit
abnormally accelerated glucose uptake and consumption (7). Glucose metabolism is considered to be a
promising target for the treatment of various types of cancer
including triple negative breast cancer (8,9).
MicroRNAs (miRNAs or miRs) are a group of small non-coding RNA
molecules, composed of ~22 nucleotides that participate in
post-transcriptional gene expression regulation and RNA silencing
(10). miRNAs interact with glucose
metabolism pathways and serve roles in cancer biology (11). miRNA-34a is closely associated with a
variety of cancer models, including breast cancer (12). The present study aimed to investigate
the interaction between miR-34a and glucose transporter 1 (GLUT1)
in triple-negative breast cancer.
Materials and methods
Patients
A total of 137 female patients were diagnosed with
triple negative breast cancer and treated at The First Affiliated
Hospital of Nanjing Medical University (Nanjing, China) from
January 2014 to January 2018. Among these, 78 patients (age range,
27–68 years; mean age, 47.1±4.7 years) were included in the current
study based on the following inclusion and exclusion criteria.
Inclusion criteria: i) Patients pathologically diagnosed with
triple negative breast cancer; ii) patients with no prior diagnosis
or treatment; iii) an education level sufficient to understand the
study procedure (high school diploma or above). Exclusion criteria:
i) Patients diagnosed with other malignancies; ii) patients who
received treatment prior to admission; iii) other breast diseases,
such as benign breast tumors. In addition, 66 healthy female
patients (age range, 25–70 years; mean age, 48.4±5.2 years) were
enrolled from January 2014 to January 2018 in the current study at
the aforementioned hospital to serve as the control group. No
significant differences were observed in age, smoking, drinking
habits or education levels between patients with breast cancer and
the control group (Table I). The
current study was approved by the Ethics Committee of the First
Affiliated Hospital of Nanjing Medical University (Nanjing, China).
Written informed consent was obtained from all participants.
 | Table I.Association of breast tissue
microRNA-34a expression and clinicopathological data. |
Table I.
Association of breast tissue
microRNA-34a expression and clinicopathological data.
| Variables | Breast cancer | Control |
|---|
| Cases | 78 | 66 |
| Age (years) | 47.1±4.7 | 48.4±5.2 |
| BMI | 23.8±1.1a | 21.0±1.2 |
| Education level |
| College
or above | 34 | 30 |
| Below
college | 44 | 36 |
| Habits |
|
Smoking | 23 (29.5%) | 19 (28.8%) |
|
Drinking | 28 (35.9%) | 25 (37.9%) |
Specimen collection
Breast tissue (100–200 mg) was collected during
biopsies from patients with triple negative breast cancer and
healthy controls. Healthy controls received breast biopsies to
detect potential breast lesions but were classified as controls
following pathological examination. In addition, blood samples (10
ml) were collected from each participant on the day of admission.
Blood samples were kept at room temperature for 2 h and serum
samples were obtained following centrifugation at 1,000 × g at room
temperature for 30 min. Tissue and serum samples were stored in
liquid nitrogen until further use.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from serum, biopsies and
in vitro cultivated cells using a TaqMan miRNA Isolation kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Total RNA was
reverse transcribed into cDNA using reactions containing 10 ng of
total RNA, 50 nmol/l stem-loop RT primer, 1X RT buffer, 0.25 mmol/l
of each dNTP, 5 U MultiScribe RT and 0.5 U RNase inhibitor
(Sigma-Aldrich; Merck KGaA). The reaction conditions were as
follows: 50°C for 30 min and 85°C for 15 min. To examine miRNA-34a
expression, qPCR was subsequently performed using the SYBR™-Green
PCR Master Mix (cat. no. 4312704; Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primer pairs for miRNA-34a (TM, 58.4°C; cat. no.
MIRAP00097-250RXN) and U6 (TM, 58.5°C; cat. no. MIRCP00001-250RXN)
were purchased from Sigma-Aldrich (Merck KGaA). The following qPCR
thermocycling conditions were used: Initial denaturation at 95°C
for 35 sec; followed by 40 cycles of 95°C for 15 sec and 62°C for
30 sec. Cq values and miRNA-34a expression were processed and
quantified using the 2−∆∆Cq method (13) and normalized to U6 small nuclear RNA.
The experiment was performed in triplicate.
Cell culture and transfection
Human triple negative breast cancer cell lines BT-20
(ATCC® HTB-19™) and MDA-MB-231 (ATCC®
HTB-26™), as well as a normal human breast epithelial tissue cell
line MCF-12F (ATCC® CRL-10783™) were purchased from the
American Type Culture Collection. All cell lines were cultured with
ATCC-formulated Eagle's Minimum Essential medium (EMEM; cat. no.
30-2003) supplemented with 10% FBS (Sigma-Aldirch; Merck KGaA) at
37°C in a 5% CO2 incubator. Cells (5×105)
were transfected with 50 nM hsa-miR-34a-3p miRNA inhibitor (cat.
no. MIH01908; Applied Biological Materials, Inc.), a miRNA
inhibitor negative control (NC) #1 (cat. no. 4464076; Thermo Fisher
Scientific, Inc.), GLUT1 small interfering RNA (siRNA;
5′-CCUCUUUGUUAAUCGCUUU-3′; Shanghai GenePharma Co., Ltd.) or a
scrambled siRNA NC 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (Shanghai
GenePharma Co., Ltd.) using the Lipofectamine 2000®
reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific,
Inc.). Transfection efficiency was confirmed using RT-qPCR prior to
subsequent experimentation, which was performed at 24 h
post-transfection. Control (C) cells were untrasnfected. NC cells
were cells transfected with miRNA inhibitor or siRNA NC.
Cell proliferation assay
After transfection and confirmation of miRNA-34a
downregulation, BT-20 and MDA-MB-231 cells were collected and mixed
with ATCC-formulated EMEM supplemented with 10% FBS to obtain a
single cell suspension with a final cell density of
4×104 cells/well. Cell suspension (0.1 ml) was added to
each well of a 96-well plate. Cells were then cultured at 37°C in a
5% CO2 incubator. Cell counting kit-8 solution (10 µl)
was added into each well after 24, 48, 72 and 96 h. Cells were then
cultured under the aforementioned conditions for a further 6 h and
a Fisherbrand™ accuSkan™ GO UV/Vis Microplate Spectrophotometer
(Thermo Fisher Scientific, Inc.) was used to measure optical
density values at 450 nm.
Glucose uptake assay
After transfection and confirmation of miRNA-34a
downregulation, BT-20 and MDA-MB-231 cells (5×105) were
washed twice with PBS and 2 ml of Krebs-Ringer-HEPES (KRH) buffer
(120 mM NaCl; 25 mM HEPES; pH 7.4; 1.2 mM MgSO4; 1.3 mM
CaCl2; 5 mM KCl and 1.3 mM KH2PO4)
containing 1 mCi of (3H)-2-deoxyglucose (PerkinElmer, Inc.). Cells
were incubated at 37°C for 25 min to initiate glucose uptake.
Glucose uptake was then halted by washing twice with ice-cold KRH
buffer. Radioactivity was measured using liquid scintillation
spectrometry (PerkinElmer, Inc.). Disintegrations per minute were
used to represent [3H]-2-deoxyglucose content in cells.
Western blot analysis
Total protein was extracted from BT-20 and
MDA-MB-231 cells using RIPA assay buffer (Thermo Fisher Scientific,
Inc.), after which total protein was quantified using a
bicinchoninic acid assay. Protein (30 µg) was loaded onto 12%
SDS-PAGE gel and transferred to PVDF membranes. Non-fat milk (5%)
was used for blocking at room temperature for 2 h. The following
primary antibodies were then incubated at 4°C for 18 h: Rabbit
anti-human GLUT1 (1:1,500; cat. no. ab15309; Abcam) and rabbit
anti-human GAPDH (1:1,300; cat. no. ab8245; Abcam). Horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibodies
(1:1,000; cat. no. MBS435036; MyBioSource, Inc.) were subsequently
used incubated at 24°C for 2 h. A Pierce ECL Western Blotting
Substrate (Thermo Fisher Scientific, Inc.) was used to produce
signals and ImageJ v1.48 software (National Institutes of Health)
was used to process data.
Statistical analysis
All experiments were performed in triplicate.
SPSS19.0 (IBM Corp.) was used for all statistical analyses.
Patients were divided into high and low expression groups according
to median breast biopsy (1.78) and serum (1.96) miRNA-34a
expression. Analysis between miRNA-34a expression and
clinicopathological patient data was performed using a
χ2 test. Data were presented as the mean ± standard
deviation and were compared using a Student's t-test (between two
groups) or a one-way ANOVA followed by a Least Significant
Difference test (for multiple groups). Receiver operating curve
(ROC) analysis was performed to evaluate the diagnostic value of
miRNA-34a expression. Patients with triple negative breast cancer
were considered true positive cases and healthy controls were
considered true negative cases. Area under the curve (AUC) >0.65
indicated potential diagnostic value. P<0.05 was considered to
indicate a statistically significant result. P-values were
subjected to Bonferroni correction for comparisons among multiple
groups.
Results
Comparison of miRNA-34a expression in
patients with triple negative breast cancer and healthy
controls
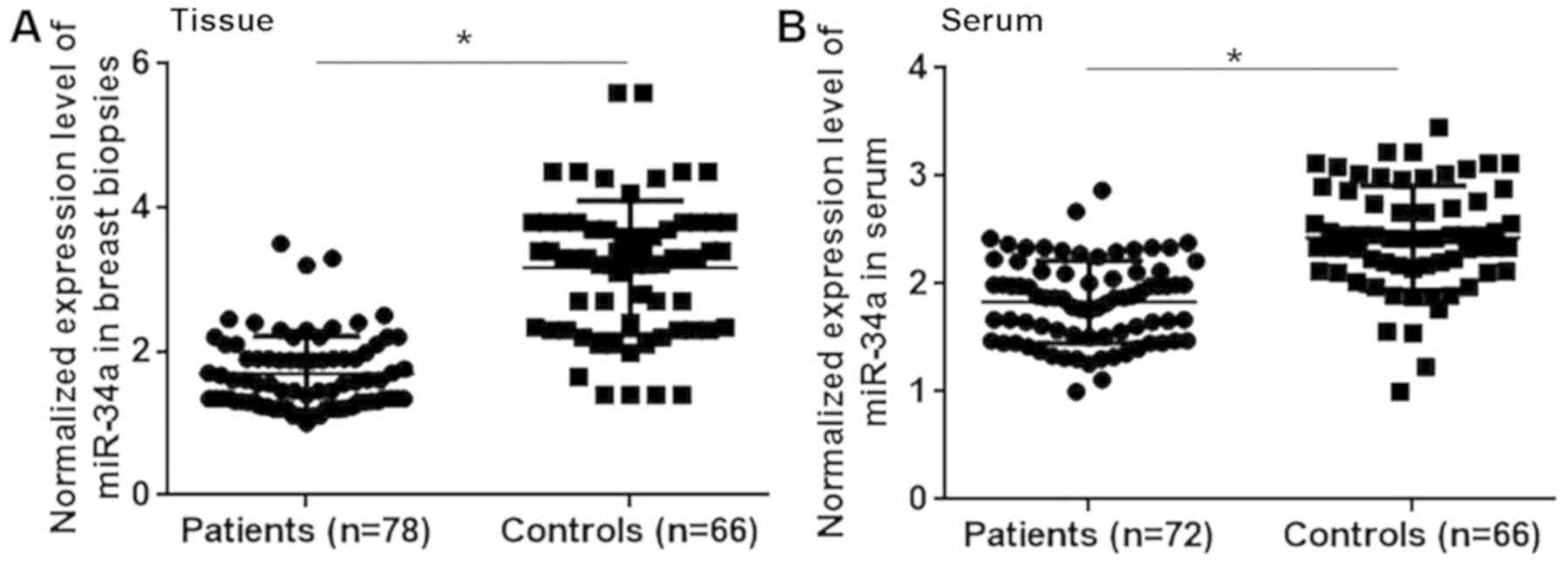
The differential expression of a gene may indicate
its involvement in a disease. Therefore, the expression of
miRNA-34a was assessed in the breast tissue and serum of patients
with triple negative breast cancer and healthy controls. The
results revealed that miRNA-34a expression was significantly lower
in patients with triple negative breast cancer compared with
healthy controls in breast tissue (Fig.
1A; P<0.05) and serum (Fig.
1B; P<0.05). Therefore, the downregulation of miRNA-34a may
participate in the pathogenesis of triple negative breast
cancer.
Diagnostic value of miRNA-34a
expression in patients with triple negative breast cancer
Receiver operating curve (ROC) analysis was
performed to evaluate the diagnostic value of miRNA-34a expression
for triple negative breast cancer. For miRNA-34a expression in
breast tissue, the AUC value was 0.9173, with a standard error of
0.02310 and a 95% confidence interval of 0.8721–0.9626
(P<0.0001; Fig. 2A). Furthermore,
the serum miRNA-34a AUC was 0.8397, with a standard error of
0.03391 and a 95% confidence interval of 0.7733–0.9062
(P<0.0001; Fig. 2B). Therefore,
miRNA-34a may be used to assist in the diagnosis of triple negative
breast cancer.
miRNA-34a expression and
clinicopathological data
Patients were divided into high and low expression
groups according to median breast biopsy and serum miRNA-34a
expression. The association between miRNA-34a expression and the
clinicopathological data of patients was analyzed using a
χ2 test. As presented in Tables II and III, miRNA-34a expression in breast tissue
and serum was not significantly associated with patient body mass
index (BMI), age, smoking and drinking habits or with the existence
of tumor distant metastasis. However, a significant association was
exhibited between miRNA-34a expression in serum (P=0.03) and biopsy
(P=0.01) and primary tumor diameter (Tables II and III).
 | Table II.Association of serum microRNA-34a
expression and clinicopathological data. |
Table II.
Association of serum microRNA-34a
expression and clinicopathological data.
| Variables | Groups | Number of cases | High expression | Low expression | χ2 | P-value |
|---|
| Age (years) | ≥45 | 42 | 19 | 23 | 0.83 | 0.36 |
|
| <45 | 36 | 20 | 16 |
|
|
| BMI | >24 | 22 | 9 | 13 | 1.03 | 0.60 |
|
| 18.5–24 | 35 | 19 | 16 |
|
|
|
| <18.5 | 21 | 11 | 10 |
|
|
| Drinking | Yes | 28 | 12 | 16 | 0.89 | 0.35 |
|
| No | 50 | 27 | 23 |
|
|
| Smoking | Yes | 26 | 12 | 14 | 0.23 | 0.63 |
|
| No | 52 | 27 | 25 |
|
|
| Primary tumor
diameter (cm) | >5 | 30 | 10 | 20 | 6.90 | 0.03 |
|
| 2–5 | 25 | 13 | 12 |
|
|
|
| <2 | 23 | 16 | 7 |
|
|
| Tumor distant
metastasis | Yes | 38 | 21 | 17 | 0.82 | 0.36 |
|
| No | 40 | 18 | 22 |
|
|
 | Table III.microRNA-34a expression in serum and
patients' clinicopathological data. |
Table III.
microRNA-34a expression in serum and
patients' clinicopathological data.
| Variables | Groups | Number of
cases | High
expression | Low
expression2 | χ2 | P-value |
|---|
| Age (years) | ≥45 | 42 | 19 | 23 | 0.83 | 0.36 |
|
| <45 | 36 | 20 | 16 |
|
|
| BMI | >24 | 22 | 8 | 14 | 2.40 | 0.30 |
|
| 18.5–24 | 35 | 20 | 15 |
|
|
|
| <18.5 | 21 | 11 | 10 |
|
|
| Drinking | Yes | 28 | 13 | 15 | 0.22 | 0.64 |
|
| No | 50 | 26 | 24 |
|
|
| Smoking | Yes | 26 | 11 | 15 | 0.92 | 0.34 |
|
| No | 52 | 28 | 24 |
|
|
| Primary tumor
diameter (cm) | >5 | 30 | 9 | 21 | 8.68 | 0.01 |
|
| 2–5 | 25 | 14 | 11 |
|
|
|
| <2 | 23 | 16 | 7 |
|
|
| Tumor distant
metastasis | Yes | 38 | 21 | 17 | 0.82 | 0.36 |
|
| No | 40 | 18 | 22 |
|
|
Effects of miRNA-34a inhibition on
glucose uptake and GLUT1 expression in cells of triple negative
breast cancer cell lines and a normal breast epithelial tissue cell
line
Data in Tables II
and III indicate that miRNA-34a
may be involved in the growth of triple negative breast cancer; a
process in which glucose metabolism serves an important role
(14). To assess the interaction
between miRNA-34a and glucose metabolism in triple negative breast
cancer, a miRNA-34a expression vector was transfected into breast
tissue cells and glucose uptake was subsequently measured. The
results revealed that miR-34a was downregulated in cancer cell
lines BT-20 and MDA-MB-231 to a greater degree than in cells of
normal breast epithelial tissue. However, the rates of
downregulation were >70% in all groups (data not shown). As
demonstrated in Fig. 3A, miRNA-34a
inhibition was significant after transfection when compared with C
and NC groups (P<0.05). Furthermore, miRNA-34a inhibition
significantly promoted glucose uptake in BT-20 and MDA-MB-231 cells
compared with the C and NC groups (P<0.05; Fig. 3B), but not in MCF-12F cells (Fig. 3B;). GLUT1 serves a pivotal role in
glucose metabolism (15). In the
current study, miRNA-34a inhibition significantly upregulated GLUT1
expression in the human triple negative breast cancer cell lines
BT-20 and MDA-MB-231 when compared with C and NC groups (P<0.05;
Fig. 3C), but not in the normal
breast epithelial tissue cell line MCF-12F (Fig. 3C).
Effect of miRNA-34a inhibition on the
proliferation of triple negative breast cancer cell lines and a
normal breast epithelial tissue cell line
An in vitro cell proliferation assay was
performed to evaluate the effects of miRNA-34a inhibition on the
proliferation of BT-20 and MDA-MB-231 cells and on a normal breast
epithelial tissue cell line, MCF-12F. As presented in Fig. 4, miRNA-34a inhibition significantly
promoted the proliferation of triple negative breast cancer cell
lines when compared with the C group (P<0.05), but had no effect
on the normal breast epithelial tissue cell line (P>0.05). In
addition, GLUT1 siRNA silencing attenuated the enhancing effects of
miRNA-34a inhibition on cancer cell proliferation (Fig. 4).
Discussion
The results of the current study demonstrated that
miRNA-34a may serve a tumor suppressive role in a triple negative
breast cancer and may participate in the development of triple
negative breast cancer, which may be achieved through the
association of miRNA-34a and glucose metabolism pathways.
Although triple negative breast cancer is considered
to be a group of heterogeneous malignancies the development of this
disease involves various genetic factors including the absence of
ER and PR and the lack of HER2 overexpression (16). A previous study has indicated that
the onset, development and progression of triple negative breast
cancer is accompanied by changes in miRNA (17). The downregulation of miRNA-34a in
breast cancer has been extensively reported in previous studies
(18). However, the expression
pattern of miRNA-34a in the triple negative subtype remains
unclear. In the present study, the expression of miRNA-34a was
demonstrated to be significantly lower in the breast tissue and
serum of patients with triple negative breast cancer compared with
healthy controls. ROC curve analysis also revealed that the
downregulation of miRNA-34a distinguished patients with triple
negative breast cancer from healthy controls. The downregulation of
miRNA-34a is therefore likely to be involved in the pathogenesis of
triple negative breast cancer.
The development of triple negative breast cancer is
complex and includes a variety of internal and external factors,
including aging, smoking, obesity and genetic factors (19–21).
Aging has been associated with favorable tumor biology (19). Smoking status and drinking habits
(20), as well as BMI beyond a
normal range (21) have also been
indicated to contribute to the development of triple negative
breast cancer. In the current study, miRNA-34a exprFession
exhibited no significant association with patient age, smoking
status, drinking habits or BMI. The results may therefore indicate
that miRNA-34a participated in the pathogenesis of triple negative
breast cancer via pathways that are independent from these patient
characteristics.
In the present study, miRNA-34a expression in breast
tissue and serum exhibited a significant association with primary
tumor diameter but not with the existence of distant tumor
metastasis. This indicated the possible involvement of miRNA-34a in
tumor growth, but not in tumor metastasis. Previous studies have
demonstrated that miRNA-34a participates in cancer cell
proliferation via interactions with glucose metabolism pathways
(22), which are critical for the
growth and survival of cancer cells (14). In the present study, miRNA-34a
inhibition significantly increased glucose uptake and upregulated
GLUT1 expression in triple negative breast cancer cells, which
serves a key role in glucose transport. miRNA-34a inhibition also
promoted the proliferation of triple negative breast cancer cells
and these inhibitory effects were attenuated by GLUT1 siRNA
silencing. miRNA-34a downregulation in triple negative cells may
therefore promote the growth of tumors by accelerating glucose
metabolism. To the best of our knowledge, the current study is the
first to examine the role of miRNA-34a in the proliferation of
breast cancer cells. The results indicated that miRNA-34a did not
significantly affect the expression of GLUT1 at mRNA level (data
not shown). Therefore, miRNA-34a may affect GLUT1 accumulation or
degradation. Future studies should assess the effects of miRNA-34a
on GLUT1 accumulation and degradation. miRNA-34a also failed to
affect other glucose transporters including GLUT2 and GLUT3 (data
not shown), indicating the specific interaction between miRNA-34a
and GLUT1 in glucose transport.
miRNA-34a inhibition indicated no significant
effects on glucose uptake or the proliferation in cell of normal
breast tissue. Therefore, miRNA-34a may serve as a potential
therapeutic marker for triple negative breast cancer.
The present study did not include miRNA-34a
overexpression experiments, which is considered to be a limitation
and should be assessed in future study. The prognostic value of
miRNA-34a for triple negative breast cancer also remains unknown.
Follow-up studies should therefore determine the prognostic value
of miRNA-34a for this disease and also assess the interactions
between miRNA-34a and BRCA1 and BRCA2, which have been revealed to
be associated with breast cancer development and progression
(5).
In conclusion, miRNA-34a expression is downregulated
in patients with triple negative breast cancer and miRNA-34a
downregulation may promote tumor growth by accelerating glucose
metabolism. The current study is challenged by a small sample size.
Future studies with a larger sample size and more cell lines are
required to support the conclusions made.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JT and ZL designed the experiments. ZL, XG and WZ
performed the experiments. JZ, LD, HL and DT collected and analyzed
data. JT drafted the manuscript. All authors approved the
manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of the First Affiliated Hospital of Nanjing Medical
University (Nanjing, China). Written informed consent was obtained
from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howlader N, Altekruse SF, Li CI, Chen VW,
Clarke CA, Ries LA and Cronin KA: US incidence of breast cancer
subtypes defined by joint hormone receptor and HER2 status. J Natl
Cancer Inst. 106:dju0552014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W, Wan YW, Allen GI, Pang K,
Anderson ML and Liu Z: Molecular pathway identification using
biological network-regularized logistic models. BMC Genomics. 14
(Suppl 8):S72013. View Article : Google Scholar
|
|
5
|
Cetin I and Topcul M: Triple negative
breast cancer. Asian Pac J Cancer Prev. 15:2427–2431. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hay N: Reprogramming glucose metabolism in
cancer: Can it be exploited for cancer therapy? Nat Rev Cancer.
16:635–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamanaka RB and Chandel NS: Targeting
glucose metabolism for cancer therapy. J Exp Med. 209:211–215.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wahdan-Alaswad RS, Edgerton SM, Salem HS
and Thor AD: Metformin targets glucose metabolism in triple
negative breast cancer. J Oncol Transl Res. 4:1292018.PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z,
Yuan Q, Zhao X, Xu N and Liang S: MicroRNA-26a regulates glucose
metabolism by direct targeting PDHX in colorectal cancer cells. BMC
Cancer. 14:4432014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
DeBerardinis RJ, Sayed N, Ditsworth D and
Thompson CB: Brick by brick: Metabolism and tumor cell growth. Curr
Opin Genet Dev. 18:54–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fidler TP, Campbell RA, Funari T, Dunne N,
Balderas Angeles E, Middleton EA, Chaudhuri D, Weyrich AS and Abel
ED: Deletion of GLUT1 and GLUT3 reveals multiple roles for glucose
metabolism in platelet and megakaryocyte function. Cell Rep.
20:881–894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W,
Chang G, Li X, Li Q, Wang S and Wang W: MicroRNA profiling implies
new markers of chemoresistance of triple-negative breast cancer.
PLoS One. 9:e962282014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: MiR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aapro M and Wildiers H: Triple-negative
breast cancer in the older population. Ann Oncol. 23 (Suppl
6):vi52–vi55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kabat GC, Kim M, Phipps AI, Li CI, Messina
CR, Wactawski-Wende J, Kuller L, Simon MS, Yasmeen S,
Wassertheil-Smoller S and Rohan TE: Smoking and alcohol consumption
in relation to risk of triple-negative breast cancer in a cohort of
postmenopausal women. Cancer Causes Control. 22:775–783. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barba M, Vici P, Pizzuti L, Di Lauro L,
Sergi D, Di Benedetto A, Ercolani C, Sperati F, Terrenato I, Botti
C, et al: Body mass index modifies the relationship between γ-H2AX,
a DNA damage biomarker, and pathological complete response in
triple-negative breast cancer. BMC Cancer. 17:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim HR, Roe JS, Lee JE, Cho EJ and Youn
HD: p53 regulates glucose metabolism by miR-34a. Biochem Biophys
Res Commun. 437:225–231. 2013. View Article : Google Scholar : PubMed/NCBI
|