Introduction
Recent epidemiology research has revealed a
worldwide increase in non-tuberculous mycobacteria (NTM)
infections. In Japan in particular, the Mycobacterium
abscessus (M. abscessus) complex is the third most
common pathogen in pulmonary diseases caused by NTM, after the
Mycobacterium avium complex and Mycobacterium
kansasii (1). The M.
abscessus complex is categorized as rapidly growing
mycobacteria, defined by visible growth within seven days, and is
one of the most difficult pathogens to treat. Over the past decade,
the M. abscessus complex has been subclassified into three
new subspecies: M. abscessus subsp. abscessus, M.
abscessus subsp. massiliense and M. abscessus
subsp. bolletii (2).
Macrolides are the key drugs used for the treatment of M.
abscessus complex infection; however, macrolides are not always
effective or in some cases they lose effectiveness during the
course of treatment. Acquired macrolide resistance is associated
with point mutations in the rrl gene, which encodes 23S rRNA
(3). An erythromycin ribosomal
methylase, encoded by erm(41) in the M. abscessus complex,
confers inducible resistance to macrolides (4). The functionality of the
erm(41) gene differs
depending on the subspecies. Most notably, M. abscessus
subsp. massiliense has been proposed to have an incomplete
erm(41) gene, which is
associated with macrolide sensitivity. In addition, some M.
abscessus subsp. abscessus strains have substitutions in
the erm(41) gene that also
lead to macrolide susceptibility. Thus, it is important to
distinguish the three kinds of subspecies and to analyze the
sequences of the rrl and erm(41) genes. Matrix-assisted laser desorption
ionization-time of flight mass spectrometry (MALDI-TOF MS) has been
used for microbial identification in recent years, and several
researchers have attempted to apply this tool to differentiate the
subspecies of the M. abscessus complex. However, different
diagnostic criteria have been used at different institutions and
the results of the method are inconsistent (5–10).
Few studies have investigated the ratio of
subspecies of the M. abscessus complex in Japan, or examined
their macrolide resistance genes (11). It is likely that regional differences
in the ratios of the subspecies and the clinical features of such
isolates may exist. In the present study, we aimed to examine the
sequence of the erm(41) gene
in M. abscessus complex subspecies. We also compared the
efficacy of using molecular testing and mass spectrometry to
classify subspecies of the M. abscessus complex.
Materials and methods
Samples and data collection
Fourteen strains of the M. abscessus complex
were obtained from each patient between July 2016 and April 2018 at
Showa University Hospital (Tokyo) or at Showa University Fujigaoka
Hospital (Yokohama). For reference, one strain of Mycobacterium
fortuitum (M. fortuitum) was collected during the
period. All strains were of sputum origin except for one M.
abscessus complex isolate from a bronchoscopy. Clinical
isolates were cultured in mycobacteria growth indicator tubes
(MGIT) and in 2% Ogawa solid medium. M. abscessus complex
and M. fortuitum were distinguished by DNA-DNA
hybridization. All clinical data were collected from medical
records. Official approval for the study was obtained in advance
from the Ethics Committee for Research at Showa University
(approved numbers 371 and 2016127). Informed consent was waived
because of the retrospective nature of the study.
Molecular testing
DNA was extracted from mycobacterial clinical
isolates using InstaGene matrix (Bio-Rad Laboratories) and stored
at −20°C. The amount of DNA extracted ranged from 104 to 452 ng/µl.
Primers for nucleic acid amplification were designed as indicated
in Table I. PCRs were performed to
amplify mutation hot spot regions in the housekeeping genes
hsp65, rpoB and ITS to classify the strains into the three
subspecies using a Mycycler ver.10.65 thermal cycler (Bio-Rad
Laboratories). The rrl and erm(41) genes were also amplified in a similar
manner. All PCR assays were carried out in 25-µl volumes containing
200 ng of template DNA, 0.1 units of Taq DNA polymerase (Roche
Diagnostics), 25 pmol of each primer, and 10 nmol of dNTPs. Cycling
parameters were 30 sec at 95°C, 30 sec at 60°C, and 60 sec at 72°C
for 30 cycles. PCR products were separated on a 5% polyacrylamide
gel or 1% agarose gel. The gels were stained with ethidium bromide
and photographed under UV illumination. The PCR products were
purified and were directly sequenced using a BigDye terminator kit
and ABI Prism 3130 xl (Applied Biosystems). When sequences
could not be obtained by direct sequencing, the PCR products were
ligated into a pGEM T easy vector (Promega), which was then used to
transform JM109 cells, as reported previously (12). Multiple clones were selected and
plasmid DNA was purified from each and sequenced. The reference
sequences for each gene were obtained from GenBank (accession
numbers CU458896.1: M. abscessus subsp. abscessus,
AP_014547.1: M. abscessus subsp. massiliense.).
 | Table I.Primer design. |
Table I.
Primer design.
| Target | Primers | Sequence | bp |
|---|
| hsp65 | hsp65F |
5′-ACCAACGATGGTGTGTCCAT-3′ | 441 |
|
| hsp65R |
5′-CTTGTCGAACCGCATACCCT-3′ |
|
| rpoB | rpoBF |
5′-GAGGGTCAGACCACGATGAC-3′ | 408 |
|
| rpoBR |
5′-AGCCGATCAGACCGATGTT-3′ |
|
| ITS | ITSF |
5′-TTGTACACACCGCCCGTC-3′ | 490 |
|
| ITS336R |
5′-CTTCTAGTGCCAAGGCATTCACC-3′ |
|
| rrl | rrl2145F |
5′-GCGAAATTCCTTGTCGGGTAAGT-3′ | 283 |
|
| rrl2427R |
5′-GGATATACGGTCCGAGGTTAG-3′ |
|
| erm(41) | erm-86F |
5′-GACCGGGGCCTTCTTCGTGAT-3′ | 673 |
|
| erm64R |
5′-GACTTCCCCGCACCGATTCC-3′ |
|
Antibiotic susceptibility test
Minimum inhibitory concentrations (MICs) of amikacin
and clarithromycin were determined by the broth microdilution
method and were interpreted according to the Clinical and
Laboratory Standards Institute document M24-A2 (13). Briefly, an appropriate volume of the
culture was transferred into 3 ml of sterilized saline until the
turbidity matched that of a 0.5 McFarland standard. A 10 µl aliquot
of the suspension was used to inoculate 11 ml of cation-adjusted
Mueller-Hinton medium and 100 µl was distributed into each of the
96 well panels. The panels were incubated for 72 h at 30°C, and
growth was determined. To test for inducible resistance to
clarithromycin, the MICs for clarithromycin were also determined
after 7 and 14 days of incubation.
Mass spectrometry
Colonies were transferred into microcentrifuge tubes
containing 300 µl of sterile deionized water, and the tubes were
incubated for 30 min at 95°C. Then samples were mixed with 900 µl
of 70% ethanol by vortexing for 1 min. The suspensions were
centrifuged at 13,000 rpm for 2 min, and the pellets were dried for
5 min at room temperature and resuspended in 20 µl of 100%
acetonitrile with zirconia beads. The mixtures were vortexed for 1
min. The samples were then suspended with 20 µl of 70% formic acid
and centrifuged at 13,000 rpm for 2 min. Subsequently, 1 µl of the
supernatant from each extract was spotted on a target plate. After
drying, 1 µl of matrix solution (saturated
α-cyano-4-hydroxycinnamic acid in 47.5% acetonitrile and 2.5%
trifluoroacetic acid) was added onto each spot. Mass spectra were
obtained on a MALDI Biotyper ver 4.0 configured with Micro flex
LT/SH with Mycobacteria Library ver.5.0 (Bruker Daltonik). Spectra
were analyzed by Flex Analysis software 3.4 and MBT compass explore
ver 4.1 (Bruker Daltonik).
Statistical analysis
The significance in each group was evaluated with
Fisher's exact test or Pearson's Chi-square test, unpaired
student's t-test, and the nonparametric Mann-Whitney test on ranks.
P<.05 was considered significant. All analyses were performed
using JMP 13.0 software (SAS Institute).
Results
Determination of M. abscessus complex
subspecies by sequencing housekeeping genes
The results of sequence analyses of housekeeping
genes are shown in Table II. To
distinguish the three subspecies, hsp65, rpoB and ITS
sequences were determined by direct sequencing and compared to
reference sequences. The sequences of the hsp65 genes from
eight strains were consistent with the reference sequence from
M. abscessus subsp. abscessus, while those from six
strains were consistent with the hsp65 reference sequence
from M. abscessus subsp. massiliense, with the
exception of one strain (no. 9626), which had a change at position
280T>A. High heterogeneity of rpoB in the M.
abscessus complex has been reported (14). The rpoB genes from eight
strains were identical to the reference gene from M.
abscessus subsp. abscessus, while a 37C>T change was
present in two strains (no. 71740 and no. 9614), and two changes
(52C>T and 391C>T) were found in another strain (no. 8548).
Six strains had rpoB sequences identical to the reference
sequence from M. abscessus subsp. massiliense, with
the exception of one substitution, 316T>C that was detected in
four strains (nos. 74369, 77944, 9626, and 9388). No amino acid
changes resulted from these nucleotide sequence differences.
Together, eight strains were identified as M. abscessus
subsp. abscessus, and six strains as M. abscessus
subsp. massiliense. The results of sequence analyses of the
ITS region were consistent with these findings. However, a novel
insertion sequence (180_181GTTGT) was found in one strain of M.
abscessus subsp. abscessus (no. 71740).
 | Table II.Sequence differences in clinical
isolates of the M. abscessus complex. |
Table II.
Sequence differences in clinical
isolates of the M. abscessus complex.
|
|
hsp65a |
rpoBb | ITSc |
|---|
|
|
|
|
|
|---|
| Strain number | 115 | 118 | 127 | 190 | 280 | 340 | 10 | 31 | 37 | 52 | 88 | 124 | 127 | 136 | 202 | 277 | 316 | 343 | 376 | 379 | 391 | 25 | 60 | 98 | 180 | 276 | Insertion |
|---|
|
CU458896d | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
9016 | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
8377 | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
9944 | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
71740 | T | T | C | C | T | C | T | T | T | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | T | G |
c.180_181insGTTGT |
|
9614 | T | T | C | C | T | C | T | T | T | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
9854 | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
8548 | T | T | C | C | T | C | T | T | C | T | T | G | C | T | C | C | T | C | C | C | T | T | A | – | C | G |
|
|
9419 | T | T | C | C | T | C | T | T | C | C | T | G | C | T | C | C | T | C | C | C | C | T | A | – | C | G |
|
|
74369 | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | C | T | T | T | C | C | G | C | T | A |
|
|
9835 | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | T | T | T | T | C | T | G | C | C | A |
|
|
77944 | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | C | T | T | T | C | C | G | C | T | A |
|
|
9626 | G | C | T | T | A | T | C | C | C | C | C | A | T | C | G | T | C | T | T | T | C | T | G | C | C | A |
|
|
9388 | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | C | T | T | T | C | C | G | C | T | A |
|
|
8006 | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | T | T | T | T | C | T | G | C | C | A |
|
|
AP014547e | G | C | T | T | T | T | C | C | C | C | C | A | T | C | G | T | T | T | T | T | C | T | G | C | C | A |
|
Patients and characteristics
As mentioned above, results were obtained for all
patients, 57% (8 of 14) of whom were infected with M.
abscessus subsp. abscessus, and 43% (6 of 14) of whom
were infected with M. abscessus subsp. massiliense.
None were infected with M. abscessus subsp. bolletii.
Table III shows the patient
characteristics. There were seven males and seven females whose
ages at diagnosis ranged from 30 to 83 years: Thirteen were
Japanese and one was Indian. According to the guidelines published
by the American Thoracic Society/Infectious Diseases Society of
America (15), all patients were
newly diagnosed with M. abscessus complex pulmonary disease,
based on at least two positive culture results derived from
pulmonary samples. As shown in Table
III, there was no significant association of the subspecies
with age, body-mass index, sex, smoking history, radiological
findings, hemoptysis, sputum smear, or C-reactive protein.
 | Table III.Characteristics of patients. |
Table III.
Characteristics of patients.
|
Characteristics | M. abscessus
subsp. abscessus (n=8) | M. abscessus
subsp. massiliense (n=6) | P-value |
|---|
| Age (years) | 69.1±18.1 | 62.5±9.7 | 0.435 |
| BMI,
kg/m2 | 19.6±3.4 | 18.6±1.8 | 0.552 |
| Sex |
|
Male | 3 (21.4) | 4 (28.5) | 0.592 |
|
Female | 5 (35.7) | 2 (14.2) |
|
| Smoking |
|
Never | 6 (42.8) | 2 (14.2) | 0.277 |
|
Ever | 2 (14.2) | 4 (28.5) |
|
| Radiological
findings |
|
Cavity | 1 (7.1) | 2 (14.2) | 0.538 |
| Symptom of
hemoptysis |
|
Yes | 2 (14.2) | 3 (21.4) | 0.58 |
| No | 6 (42.8) | 3 (21.4) |
|
| Positive smear |
|
Yes | 6 (42.8) | 3 (21.4) | 0.58 |
| No | 2 (14.2) | 3 (21.4) |
|
| Laboratory
findings |
| CRP,
mg/dl | 0.96±1.22 | 1.68±2.17 | 0.492 |
Gene status of rrl and erm(41)
Sequence differences identified in the rrl
and erm(41) genes are
summarized in Table IV. In the
rrl gene, a A>G change was detected at position 2059 in
one strain (no. 8006), but no other alterations were found. As for
the erm(41) gene,
nucleotides at positions 64_65 and 159_432 were deleted in strains
of M. abscessus subsp. massiliense, compared to the
M. abscessus subsp. abscessus strains. Eight
substitutions were found in the M. abscessus subsp.
abscessus isolates, whereas no substitutions were found in
the strains of M. abscessus subsp. massiliense. In
those isolates of M. abscessus subsp. abscessus,
28T>C, 238A>G and 419C>T substitutions were responsible
for the amino-acid changes W10R, I80V and P140L, respectively. As
shown in Fig. 1, the sizes of the
PCR products amplified from the erm(41) genes were consistent with sequencing
results (673 base pairs for M. abscessus subsp.
abscessus, and 397 base pairs for M. abscessus subsp.
massiliense), and identifications based on the size of the
erm(41) gene were consistent
with those based on hsp65, rpoB and ITS sequences.
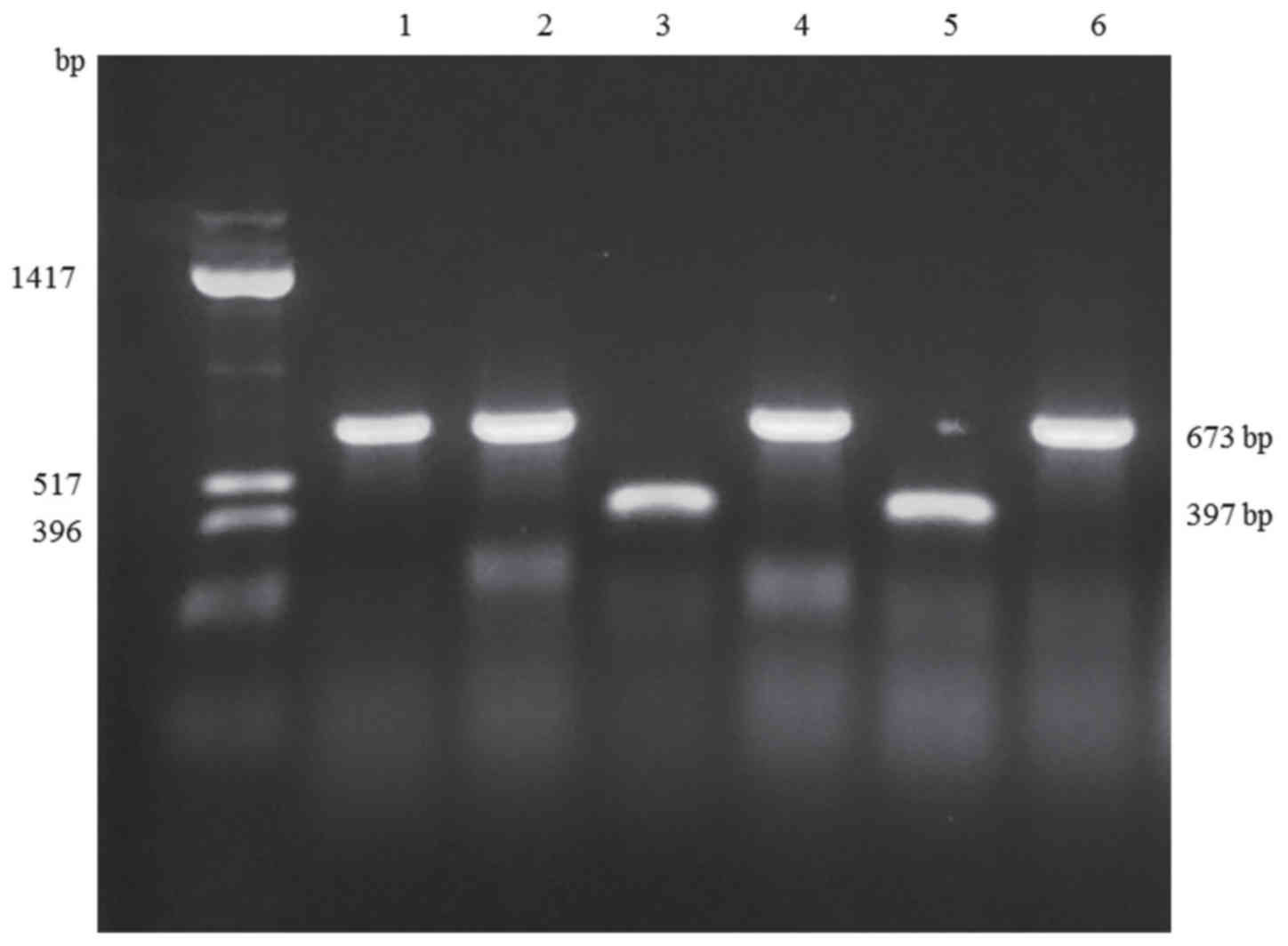 | Figure 1.Representative PCR products for the
erm(41) gene. The amplified
products from M. abscessus subsp. abscessus strains
(no. 9016, no. 8377, no. 9944 and no. 9854) were 673 bp in length,
whereas those from M. abscessus subsp. massiliense
strains (no. 9835 and no. 9626) were 397 bp in length. Far left
lane, DNA size standard; Lane 1, no. 8377; Lane 2, 9016; Lane 3,
9626; Lane 4, 9944; Lane 5, 9835; Lane 6, 9854. |
 | Table IV.Sequence differences in the
rrl and erm(41) genes
from clinical isolates of the M. abscessus complex. |
Table IV.
Sequence differences in the
rrl and erm(41) genes
from clinical isolates of the M. abscessus complex.
|
|
rrla |
erm(41)b |
|---|
|
|
|
|
|---|
| Strain number | 2058 | 2059 | −28 | −4 | 28 | 41 | 46 | 64 | 65 | 85 | 90 | 109 | 120 | 123 | 159 | 238 | 255 | 279 | 330 | 336 | 419 | 432 | 438 | 466 | Amino acid
change |
|---|
|
CU458896c | A | A | A | C | T | C | A | C | G | G | C | G | A | A | T | A | G | G | A | T | C | G | A | G |
|
|
|
|
9016 | A | A | A | C | T | C | A | C | G | G | C | G | A | A | C | Gf | G | G | C | T | C | G | A | G |
| I80Vf |
|
|
8377 | A | A | A | C | T | C | A | C | G | G | C | G | A | A | T | A | G | G | A | T | C | G | A | G |
|
|
|
|
9944 | A | A | A | C | Ce | C | A | C | G | G | C | G | A | A | C | Gf | G | G | C | T | C | G | A | G | W10Re | I80Vf |
|
|
71740 | A | A | A | C | T | C | A | C | G | G | C | G | A | A | C | Gf | A | T | C | C | C | G | A | G |
| I80Vf |
|
|
9614 | A | A | A | C | T | C | A | C | G | G | C | G | G | A | C | Gf | A | T | C | C | C | G | A | G |
| I80Vf |
|
|
9854 | A | A | A | C | T | C | A | C | G | G | C | G | A | A | T | A | G | G | A | T | C | G | A | G |
|
|
|
|
8548 | A | A | A | C | T | C | A | C | G | G | C | G | A | A | C | Gf | A | T | C | C | Tg | G | A | G |
| I80Vf | P140Lg |
|
9419 | A | A | A | C | T | C | A | G | G | G | C | G | A | A | C | Gf | A | T | C | C | C | G | A | G |
|
I80Vf |
|
|
74369 | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
9835 | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
77944 | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
9626 | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
9388 | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
8006 | A | G | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
|
AP014547d | A | A | G | T | T | A | G | – | – | T | T | A | A | G | – | – | – | – | – | – | – | – | C | A |
|
|
|
Antimicrobial sensitivity
Table V shows the
antibiotic susceptibility of the M. abscessus strains to
amikacin and clarithromycin. The MICs of amikacin ranged from 2 to
16 µg/ml, which indicated that all strains were sensitive. There
was no difference in the MICs between the two subspecies. M.
abscessus subsp. abscessus isolates were sensitive to
clarithromycin on day 3, but the MICs were significantly higher on
day 14 with one exception (no. 9944). In contrast, the strains of
M. abscessus subsp. massiliense were susceptible to
clarithromycin on days 3 through 14, except for one strain (no.
8006), which showed resistance from the start. Strain no. 9626 was
sensitive early in the testing period, but the MIC was about 4-fold
higher on day 14.
 | Table V.Antibiotic susceptibilities of M.
abscessus complex isolates based on MIC (µg/ml) values. |
Table V.
Antibiotic susceptibilities of M.
abscessus complex isolates based on MIC (µg/ml) values.
|
|
| Amikacin | Clarithromycin |
|---|
|
|
|
|
|
|---|
| Isolate | Strains no. | Day 3 | Day 3 | Day 7 | Day 14 |
|---|
| M. abscessus
subsp. abscessus |
9016 | 16 | 0.125 | 64 | 64 |
|
|
8377 | 16 | 0.5 | 64 | >128 |
|
|
9944 | 8 | ≤0.06 | ≤0.06 | ≤0.06 |
|
| 71740 | 8 | 0.25 | 32 | 32 |
|
|
9614 | 8 | 0.25 | 64 | 64 |
|
|
9854 | 8 | 0.125 | >128 | >128 |
|
|
8548 | 8 | ≤0.06 | 64 | >128 |
|
|
9419 | 8 | 0.5 | 32 | 32 |
| M. abscessus
subsp. massiliense | 74369 | 16 | 0.25 | 0.25 | 0.25 |
|
|
9835 | 16 | ≤0.06 | ≤0.06 | ≤0.06 |
|
| 77944 | 4 | ≤0.06 | ≤0.06 | ≤0.06 |
|
|
9626 | 16 | 0.125 | 0.25 | 0.5 |
|
|
9388 | 8 | ≤0.06 | ≤0.06 | ≤0.06 |
|
|
8006 | 2 | >128 | >128 | >128 |
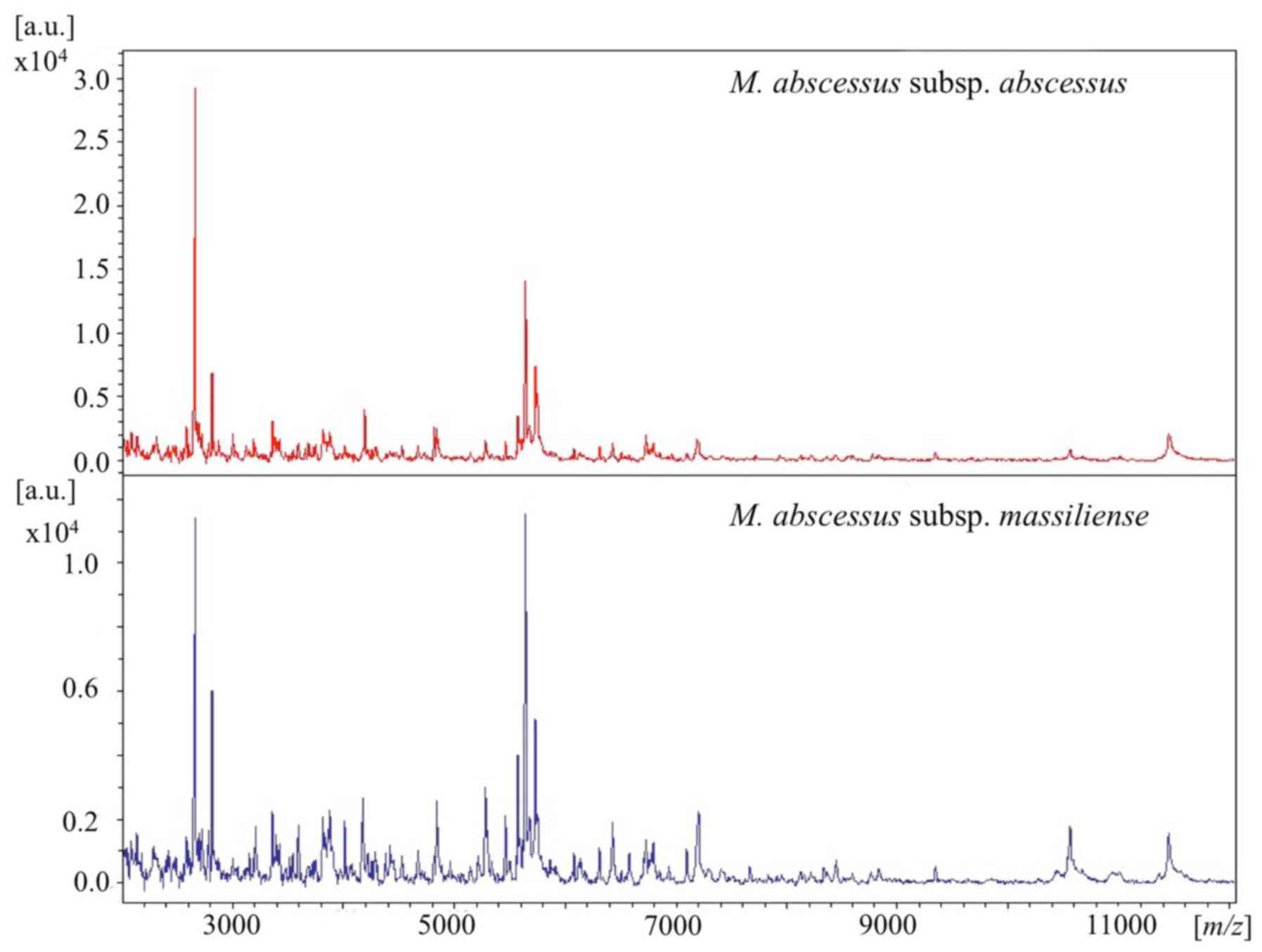
MALDI-TOF MS analysis
The details of mass spectra are shown in Figs. 2 and 3. Out of the 14 isolates, 11 were analyzed
using MALDI-TOF MS. For reference, one clinical isolate of M.
fortuitum was simultaneously analyzed in a similar manner. The
11 strains were identified as M. abscessus complex (score
range, 1.66 to 2.14) and were correctly differentiated from the
strain of M. fortuitum (Fig.
S1). As shown in Fig. 2, the
representative spectra of M. abscessus complex subspecies
were similar at a laser frequency of 50 Hz across 2,000 to 12,000
m/z. When magnifying the spectrum (Fig. 3), distinctive peaks reported
previously (8,10) were detected in some cases, but not in
all samples. More specifically, peaks around 4390, 7639, 8781 and
9473 m/z for M. abscessus subsp. abscessus and
peaks around 4385, 7669, and 8767 m/z for M.
abscessus subsp. massiliense were found. However, each
baseline was unstable and the peaks were wide and low for these
strains. Moreover, discriminating peaks were mostly of low
intensity or were overlapping. Thus, detection of clearly
identifiable differences between strains of different subspecies
was extremely difficult. Overall, it was possible to discriminate
between M. abscessus subsp. abscessus and M.
abscessus subsp. massiliense by using mass spectra only
in some cases.
Discussion
In the current study, M. abscessus complex
isolates obtained from patients treated at our institutes located
in the Tokyo-Yokohama area were analyzed. Overall, 57% (8 of 14) of
the isolates were identified as M. abscessus subsp.
abscessus, 43% (6 of 14) as M. abscessus subsp.
massiliense, and none as M. abscessus subsp.
bolletii. It has been reported that M. abscessus
subsp. abscessus is the most predominant subspecies of the
complex followed by M. abscessus subsp. massiliense,
and that M. abscessus subsp. bolletii is quite rare,
ranging from 0–3% in Japan (11,16–19).
There were no significant differences in clinical features between
M. abscessus subsp. abscessus and M. abscessus
subsp. massiliense. Clinical characteristics of the M.
abscessus complex did not help us to distinguish those
subspecies, which is consistent with findings reported in the
literature (11,20).
The proportion of the M. abscessus complex
subspecies varies depending on the region from which they are
isolated. Compared to Western Europe, the prevalence of M.
abscessus subsp. bolletii is lower in East Asian
countries (21–24). Although, M. abscessus subsp.
abscessus is the most predominant subspecies of the complex
in most parts of the world, some studies have reported that M.
abscessus subsp. massiliense is more abundant than M.
abscessus subsp. abscessus in some parts of East Asia
such as Taiwan and Korea (5,21,22,25).
Actually whole-genome sequencing analyses revealed genetic
distinctions between M. abscessus subsp. abscessus
isolates in Asia and Western Europe (26), suggesting that the phylogenetic
diversity correlates with the regional ratio of the subspecies.
The current commercial system for NTM
differentiation in Japan consists of the DNA-DNA hybridization
method, which is unable to differentiate subspecies of the M.
abscessus complex. The three subspecies also cannot be
distinguished by 16S rRNA gene sequencing, which is commonly used
for bacterial taxonomy in academic research, because the 16S rRNA
genes in the three subspecies are 100% identical (27). The differentiation requires
sequencing of several housekeeping genes, which is not easy to
accomplish in most mycobacteriology laboratories. Hence, these
three subspecies have not been distinguished in hospital
laboratories. Sequencing of a single target gene may lead to
inaccurate identification of closely related subspecies; however,
multilocus sequence analyses of the M. abscessus complex
have been described using hsp65, rpoB, ITS, gyrB, dnaA,
recA and secA (28).
Although some other methods based on technology developed by
multilocus sequence analyses have been designed, such as
variable-number tandem repeat analysis (18,19,29,30) and
multiplex PCR (17,31), those methods are complicated. In the
present study, subspecies of the M. abscessus complex were
differentiated based on partial sequences of the hsp65 and
rpoB genes, and results of ITS sequencing were also
consistent in differentiating the two subspecies. In a subset of
M. abscessus complex isolates, a hybrid genetic pattern for
the hsp65 and rpoB genes has been reported (32,33),
presumably the result of horizontal gene transfer between the
subspecies. In such cases ITS gene analysis was essential to
identify the subspecies. Sequencing of at least three housekeeping
genes should therefore be carried out for subspecies
identification.
Numerous institutions are seeking an alternative way
to distinguish the M. abscessus subspecies in clinical
practice. MALDI-TOF MS has been evaluated for the identification of
microorganisms including mycobacteria. However in this study, it
was not possible to distinguish between the M. abscessus
subspecies of all isolates by MALDI-TOF MS. Although several
institutes have reported the efficacy of MALDI-TOF MS in
differentiating the three subspecies of the M. abscessus
complex (5–10), methods for sample preparation and
analysis, and diagnostic criteria have not been standardized. When
defining the range from 2,000 to 20,000 m/z, ribosomal
protein accounts for 50 to 70% of the peptide detected by MALDI-TOF
MS. Thus, it is easy to distinguish between species that contain
diverse ribosomal proteins. In fact, the current study revealed
obvious differences between M. fortuitum and the M.
abscessus complex by mass spectra (Fig. S1). However, the mass spectrometry
peaks may be affected by variations in culture media formulations,
duration of growth, and other conditions. The utility of MGIT
liquid medium (10), 5% sheep blood
agar (6), Middlebrook 7H11 (7), and Lowenstein-Jensen agar (9) have been reported for MALDI-TOF MS. In
the present study, samples were prepared from colonies incubated on
2% Ogawa solid medium. Thus, the diagnostic criteria for MALDI-TOF
analysis differ depending on the laboratory carrying out the
analyses. Since there are few differences in the ribosomal proteins
between the M. abscessus complex subspecies, and the sample
preparation for mass spectra is even affected by climate, the
procedure is often poorly reproducible. Considerable effort would
be necessary to optimize and standardize a protocol to obtain
reproducible mass spectrometry results for differentiating the
M. abscessus complex subspecies. A proteomics study revealed
specific immunogenic proteins in the M. abscessus complex;
therefore, the subspecies may be differentiated using immunoassays
(34).
Mutations in rrl gene confer acquired
resistance and nucleotide T28 of erm(41) is associated with inducible
resistance. Since most of the alterations identified in this study
were in the erm(41) gene
(92.9%; 13 of 14 isolates) rather than the rrl gene (7.1%;1
of 14), we concluded that the main cause of treatment failure for
M. abscessus complex infections was inducible resistance
encoded by the erm(41) gene.
One strain of M. abscessus subsp. massiliense (no.
8006) harboring a point mutation 2059A>G in the rrl gene
showed resistance to clarithromycin from day 3 through day 14 (MIC
> 128 µg/ml). The presence of rrl mutations was reported
in up to 30% of newly isolated strains in East Asia (16,35),
whereas no rrl mutations were identified at the time of
diagnosis in Spain (36). It has
been reported that mutations in the rrl gene rapidly
accumulate following clarithromycin use in monotherapy (3). Conversely, several studies have
reported on acquired resistance due to rrl mutations in the
absence of any macrolide exposure (16,37).
However, an in vitro study reported that rrl
mutations at position 2058 or 2059 were observed during incubation
with clarithromycin (38). In East
Asia, low dose macrolides are often administered for prolonged
periods to treat chronic respiratory disorders, such as diffuse
panbronchiolitis, to stimulate an immunomodulatory effect (39). Hence, careful attention is needed
when prescribing macrolides to patients initially diagnosed with
pulmonary disease caused by M. abscessus complex. One strain
of M. abscessus subsp. abscessus (no. 9944) carried a
substitution at position 28T>C in the erm(41) gene, which resulted in an amino acid
change (W10R), and showed a low MIC value for clarithromycin. Other
strains that harbored amino acid substitutions at I80V (238A>G)
or P140L (419C>T) showed inducible resistance. These data are in
agreement with previous reports (4,37)
suggesting that the 5′ end of the erm(41) gene is a key region because this
region is also predicted to carry a second open reading frame
encoding a leader peptide that regulates expression of the
erm(41) gene itself
(4). In M. abscessus subsp.
massiliense, two base deletions (61_62del) have been
commonly detected, in addition to longer deletions (159_432del),
which are consistent with the findings of the present study.
Strains of M. abscessus subsp. abscessus harboring a
C>T substitution at position 19, resulting in a stop codon
(R7stop), have been previously identified in the findings of a case
report (40). The data also
highlight the importance of the 5′ end of the erm(41) gene. With rare exceptions, such as the
presence of a full-length erm(41) gene (41,42),
M. abscessus subsp. massiliense is generally not
associated with inducible macrolide resistance because most strains
harbor a truncated erm(41)
gene (4,21,43).
Interestingly, in the present study, one strain (no. 9626) showed a
low initial MIC but the MIC increased 4-fold by 14 days. This
observation suggests that mechanisms not involving
erm(41) can cause inducible
resistance. No strains that simultaneously harbored both a
rrl mutation and nucleotide T28 in erm(41) were detected in this study; however,
numerous studies have revealed that both resistance mechanisms can
occur concurrently (36,38), and that a functional
erm(41) gene does not
exclude selection for rrl mutations (44). Transcriptome analysis of M.
abscessus complex revealed the presence of several novel open
reading frames, which could be activated by stressful conditions,
such as hypoxia, to persist (45).
Therefore, in addition to antimicrobial resistance genes,
pathogenetic gene alterations should also be investigated in the
future.
As described above, the two subspecies of the M.
abscessus complex were found, which is consistent with the
results of other studies showing the predominance of M.
abscessus subsp. abscessus and M. abscessus
subsp. massiliense in East Asia (11,16,21,24).
Data from the current study indicate that the principal difference
between the two subspecies was the size of the erm(41) gene. Several studies have revealed the
presence of a complete erm(41) gene in M. abscessus subsp.
massiliense strains (41,42) and
a truncated erm(41) gene in
M. abscessus subsp. bolletii (36), suggesting that M. abscessus
subsp. massiliense acquired a full-length
erm(41) gene by horizontal
transfer from M. abscessus subsp. abscessus or M.
abscessus subsp. bolletii, and that a truncated
erm(41) gene was transferred
from M. abscessus subsp. massiliense to M.
abscessus subsp. bolletii. Additionally, a change at
position 28T>C has been reported in M. abscessus subsp.
bolletii (21). Therefore,
although horizontal gene transfer between subspecies is probably
quite rare, erm(41) PCR is
not proposed as the best way to differentiate M. abscessus
complex subspecies. However, erm(41) PCR can be easily and efficiently used
for the prediction of sensitivity to clarithromycin in the M.
abscessus complex. The phenotypic function of a putative
erm(41) gene is important
for the clinician from a strictly pragmatic standpoint. Likewise,
M. abscessus complex subspecies should be categorized based
on the presence of a functional erm(41) gene and macrolide sensitivity
especially if horizontal gene transfer increases in the future.
In conclusion, the present study demonstrates the
features of M. abscessus subsp. abscessus and M.
abscessus subsp. massiliense, isolated in the
Tokyo-Yokohama area. No strains of M. abscessus subsp.
bolletii were detected. It was not possible to differentiate
the two subspecies by clinical features in pulmonary infection, and
results of mass spectrometry analysis of both these subspecies were
highly similar; however, it is possible to predict clarithromycin
susceptibility in strains of the two species by PCR amplification
of the erm(41) gene. This is
a simple and useful method that can be carried out routinely in
hospital laboratories, and is recommended to predict inducible
resistance to macrolides before determining the MICs, which
requires 14 days of incubation for M. abscessus complex
subspecies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mr. Sadahiro
Ichimura and Mr. Hidetoshi Yamamoto (BML Inc., Saitama, Japan) for
their microbiological techniques and Ms. Azumi Fujinaga (Bruker
Japan K.K., Yokohama, Japan) for help with the mass spectra
analysis.
Funding
This work was supported by Grant-in-Aid for
Scientific Research(C) (grant no. 17K09021).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
AM, FY, TF, YY and YS examined and cared for the
patients. AM, FY, TF, YY, YS and KF developed the concept, designed
the experiments and analyzed the data. AM wrote the manuscript with
contributions from all authors, who commented on it at all
stages.
Ethics approval and consent to
participate
Official approval for the study was obtained in
advance from the Ethics Committee for Research at Showa University
(approval nos. 371 and 2016127). Informed consent was waived due to
the retrospective nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Namkoong H, Kurashima A, Morimoto K,
Hoshino Y, Hasegawa N, Ato M and Mitarai S: Epidemiology of
pulmonary nontuberculous Mycobacterial disease, Japan. Emerg Infect
Dis. 22:1116–1117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee MR, Sheng WH, Hung CC, Yu CJ, Lee LN
and Hsueh PR: Mycobacterium abscessus complex infections in humans.
Emerg Infect Dis. 21:1638–1646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wallace RJ, Meier A, Brown BA, Zhang Y,
Sander P, Onyi GO and Böttger EC: Genetic basis for clarithromycin
resistance among isolates of Mycobacterium chelonae and
Mycobacterium abscessus. Antimicrob Agents Chemother.
40:1676–1681. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nash KA, Brown-Elliott AB and Wallace RJ
Jr: A Novel gene, erm(41), confers inducible macrolide resistance
to clinical isolates of Mycobacterium abscessus but is
absent from mycobacterium chelonae. Antimicrob Agents
Chemother. 53:1367–1376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Teng SH, Chen CM, Lee MR, Lee TF, Chien
KY, Teng LJ and Hsueh PR: Matrix-assisted laser desorption
ionization-time of flight mass spectrometry can accurately
differentiate between Mycobacterium masilliense (M.
abscessus subspecies bolletti) and M. abscessus
(Sensu Stricto). J Clin Microbiol. 51:3113–3116. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fangous MS, Mougari F, Gouriou S, Calvez
E, Raskine L, Cambau E, Payan C and Hery-Arnaud G: Classification
algorithm for subspecies identification within the Mycobacterium
abscessus species, based on matrix-assisted laser desorption
ionization-time of flight mass spectrometry. J Clin Microbiol.
52:3362–3369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Panagea T, Pincus DH, Grogono D, Jones M,
Bryant J, Parkhill J, Floto RA and Gilligan P: Mycobacterium
abscessus complex identification with matrix-assisted laser
desorption ionization-time of flight mass spectrometry. J Clin
Microbiol. 53:2355–2358. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki H, Yoshida S, Yoshida A, Okuzumi K,
Fukusima A and Hishinuma A: A novel cluster of Mycobacterium
abscessus complex revealed by matrix-assisted laser desorption
ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Diagn
Microbiol Infect Dis. 83:365–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Buckwalter SP, Olson SL, Connelly BJ,
Lucas BC, Rodning AA, Walchak RC, Deml SM, Wohifiel SL and
Wengenack NL: Evaluation of matrix-assisted laser desorption
ionization-time of flight mass spectrometry for identification of
Mycobacterium species, Nocardia species, and other
aerobic actinomycetes. J Clin Microbiol. 54:376–384. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kehrmann J, Wessel S, Murali R, Hampel A,
Bange FC, Buer J and Mosel F: Principal component analysis of MALDI
TOF MS mass spectra separates M. abscessus (sensu
stricto) from M. massiliense isolates. BMC Microbiol.
16:242016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harada T, Akiyama Y, Kurashima A, Nagai H,
Tsuyuguchi K, Fujii T, Yano S, Shigeto R, Kuraoka T, Kajiki A, et
al: Clinical and microbiological differences between
Mycobacterium abscessus and Mycobacterium massiliense
lung diseases. J Clin Microbiol. 50:3556–3561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukuchi K, Hagiwara T, Nakamura K,
Ichimura S, Tatsumi K and Gomi K: Identification of the regulatory
region required for ubiquitination of the cyclin kinase inhibitor,
p21. Biochem Biophys Res Commun. 293:120–125. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Woods GL, Brown-Elliott BA, Conville PS,
Desmond EP, Hall GS and Lin G: Susceptibility testing of
Mycobacteria, Nocardia and other aerobic actinomycetes; Approved
Standard-Second edition. Clin Lab Stand Inst. 26:1–61. 2011.
|
|
14
|
Macheras E, Roux AL, Bastian S, Leao SC,
Palaci M, Tardy VS, Gutierrez C, Richter E, Gerdes SR, Pfyffer G,
et al: Multilocus sequence analysis and rpoB sequencing of
Mycobacterium abscessus (sensu lato) strains. J Clin
Microbiol. 49:491–499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Griffith DE, Aksamit T, Brown-Elliott BA,
Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G,
Iademarco MF, et al: An official ATS/IDSA statement: Diagnosis,
treatment, and prevention of nontuberculous mycobacterial diseases.
Am J Respir Crit Care Med. 175:367–416. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshida S, Tsuyuguchi K, Suzuki K, Tomita
M, Okada M, Hayashi S, Iwamoto T and Saito H: Further isolation of
Mycobacterium abscessus subsp. abscessus and subsp.
bolletii in different regions of Japan and susceptibility of
these isolates to antimicrobial agents. Int J Antimicrob Agents.
42:226–231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakanaga K, Sekizuka T, Fukano H,
Sakakibara Y, Takeuchi F, Wada S, Ishii N, Makino M, Kuroda M and
Hoshino Y: Discrimination of Mycobacterium abscessus subsp.
massiliense from Mycobacterium abscessus subsp.
abscessus in clinical isolates by multiplex PCR. J Clin
Microbiol. 52:251–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshida S, Arikawa K, Tsuyuguchi K,
Kurashima A, Harada T, Nagai H, Suzuki K, Iwamoto T and Hayashi S:
Investigation of the population structure of Mycobacterium
abscessus complex strains using 17-locus variable number tandem
repeat typing and the further distinction of mycobacterium
massiliense hsp65 genotypes. J Med Microbiol. 64:254–261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kusuki M, Osawa K, Arikawa K, Tamura M,
Shigemura K, Shirakawa T, Nakamura T, Nakamachi Y, Fujisawa M,
Saegusa J and Tokimatsu I: Determination of the antimicrobial
susceptibility and molecular profile of clarithromycin resistance
in the Mycobacterium abscessus complex in Japan by variable
number tandem repeat analysis. Diagn Microbiol Infect Dis.
91:256–259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH,
Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, et al: Clinical
significance of differentiation of Mycobacterium massiliense
from Mycobacterium abscessus. Am J Respir Crit Care Med.
183:405–410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH,
Kim BJ and Kook YH: Mycobacterium massiliense is
differentiated from Mycobacterium abscessus and
mycobacterium bolletii by erythromycin ribosome
methyltransferase gene (erm) and clarithromycin susceptibility
patterns. Microbiol Immunol. 54:347–353. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim BJ, Yi SY, Shim TS, Do SY, Yu SK, Park
YG, Kook YH and Kim BJ: Discovery of a novel hsp65 genotype within
Mycobacterium massiliense associated with the rough colony
morphology. PLoS One. 7:e384202012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim SY, Kang YA, Bae IK, Yim JJ, Park MS,
Kim YS, Kim SK, Chang J and Jeong SH: Standardization of multilocus
sequence typing scheme for Mycobacterium abscessus and
Mycobacterium massiliense. Diagn Microbiol Infect Dis.
77:143–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo L, Li B, Chu H, Huang D, Zhang Z,
Zhang J, Gui T, Xu L, Zhao L, Sun X and Xiao H: Characterization of
Mycobacterium abscessus subtypes in Shanghai of China: Drug
sensitivity and bacterial epidemicity as well as clinical
manifestations. Medicine (Baltimore). 95:e23382016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koh WJ, Jeong BH, Kim SY, Jeon K, Park KU,
Jhun BW, Lee H, Park HY, Kim DH, Huh HJ, et al: Mycobacterial
characteristics and treatment outcomes in Mycobacterium
abscessus lung disease. Clin Infect Dis. 64:309–316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davidson RM, Hasan NA, Reynolds PR, Totten
S, Garcia B, Levin A, Ramamoorthy P, Heifets L, Daley CL and Strong
M: Genome sequencing of Mycobacterium abscessus isolates
from patients in the United States and comparisons to globally
diverse clinical strains. J Clin Microbiol. 52:3573–3582. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leao SC, Tortoli E, Viana-Niero C, Ueki
SY, Lima KV, Lopes ML, Yubero J, Menendez MC and Garcia MJ:
Characterization of mycobacteria from a major Brazilian outbreak
suggests that revision of the taxonomic status of members of the
Mycobacterium chelonae-M. abscessus group is needed. J Clin
Microbiol. 47:2691–2698. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tan JL, Khang TF, Ngeow YF and Choo SW: A
phylogenomic approach to bacterial subspecies classification: Proof
of concept in Mycobacterium abscessus. BMC Genomics.
14:8792013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kikuchi T, Watanabe A, Gomi K, Sakakibara
T, Nishimori K, Daito H, Fujimura S, Tazawa R, Inoue A, Ebina M, et
al: Association between mycobacterial genotypes and disease
progression in Mycobacterium avium pulmonary infection.
Thorax. 64:901–907. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wong YL, Ong CS and Ngeow YF: Molecular
typing of Mycobacterium abscessus based on tandem-repeat
polymorphism. J Clin Microbiol. 50:3084–3088. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mougari F, Raskine L, Ferroni A, Marcon E,
Sermet-Gaudelus I, Veziris N, Heym B, Gaillard JL, Nassif X and
Cambau E: Clonal relationship and differentiation among
Mycobacterium abscessus isolates as determined using the
semiautomated repetitive extragenic palindromic sequence PCR-based
diversilab system. J Clin Microbiol. 52:1969–1977. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim HY, Kook Y, Yun YJ, Park CG, Lee NY,
Shim TS, Kim BJ and Kook YH: Proportions of Mycobacterium
massiliense and Mycobacterium bolletii strains among
Korean Mycobacterium chelonae-Mycobacterium abscessus group
isolates. J Clin Microbiol. 46:3384–3390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim BJ, Kim GN, Kim BR, Shim TS, Kook YH
and Kim BJ: Phylogenetic analysis of Mycobacterium
massiliense strains having recombinant rpoB gene laterally
transferred from Mycobacterium abscessus. PLoS One.
12:e01792372017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Steindor M, Nkwouano V, Stefanski A,
Stuehler K, Loerger TR, Bogumil D, Jacobsen M, Mackenzie CR and
Kalscheuer R: A proteomics approach for the identification of
species-specific immunogenic proteins in the Mycobacterium
abscessus complex. Microbes Infect. Nov 13–2018.(Epub ahead of
print). doi: 10.1016/j.micinf.2018.10.006. PubMed/NCBI
|
|
35
|
Lee SH, Yoo HK, Kim SH, Koh WJ, Kim CK,
Park YK and Kim HJ: Detection and assessment of clarithromycin
inducible resistant strains among Korean Mycobacterium
abscessus clinical strains: PCR methods. J Clin Lab Anal.
28:409–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rubio M, March F, Garrigó M, Moreno C,
Español M and Coll P: Inducible and acquired clarithromycin
resistance in the Mycobacterium abscessus complex. PLoS One.
10:e01401662015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bastian S, Veziris N, Roux AL, Brossier F,
Gaillard JL, Jarlier V and Cambau E: Assessment of clarithromycin
susceptibility in strains belonging to the Mycobacterium
abscessus group by erm(41) and rrl sequencing. Antimicrob
Agents Chemother. 55:775–781. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mougari F, Bouziane F, Crockett F, Nessar
R, Chau F, Veziris N, Sapriel G, Raskine L and Cambau E: Selection
of resistance to clarithromycin in Mycobacterium abscessus
subspecies. Antimicrob Agents Chemother. 61(pii): e00943–16.
2016.PubMed/NCBI
|
|
39
|
Kudoh S and Keicho N: Diffuse
panbronchiolitis. Clin Chest Med. 33:297–305. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim SY, Shin SJ, Jeong BH and Koh WJ:
Successful antibiotic treatment of pulmonary disease caused by
Mycobacterium abscessus subsp. abscessus with C-to-T
mutation at position 19 in erm(41) gene: Case report. BMC Infect
Dis. 16:2072016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shallom SJ, Gardina PJ, Myers TG,
Sebastian Y, Conville P, Calhoun LB, Tettelin H, Olivier KN, Uzal
G, Sampaio EP, et al: New rapid scheme for distinguishing the
subspecies of the Mycobacterium abscessus group and
identifying Mycobacterium massiliense isolates with
inducible clarithromycin resistance. J Clin Microbiol.
51:2943–2949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brown-Elliott BA, Vasireddy S, Vasireddy
R, Iakhiaeva E, Howard ST, Nash K, Parodi N, Strong A, Gee M, Smith
T and Wallace RJ Jr: Utility of sequencing the erm(41) gene in
isolates of Mycobacterium abscessus subsp. abscessus with
low and intermediate clarithromycin MICs. J Clin Microbiol.
53:1211–1215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoshida S, Tsuyuguchi K, Suzuki K, Tomita
M, Okada M, Shimada R and Hayashi S: Rapid identification of
strains belonging to the Mycobacterium abscessus group
through erm(41) gene pyrosequencing. Diagn Microbiol Infect Dis.
79:331–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maurer FP, Rüegger V, Ritter C, Bloemberg
GV and Böttger EC: Acquisition of clarithromycin resistance
mutations in the 23S rRNA gene of Mycobacterium abscessus in
the presence of inducible erm(41). J Antimicrob Chemother.
67:2606–2611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miranda-CasoLuengo AA, Staunton PM, Dinan
AM, Lohan AJ and Loftus BJ: Functional characterization of the
Mycobacterium abscessus genome coupled with condition
specific transcriptomics reveals conserved molecular strategies for
host adaptation and persistence. BMC Genomics. 17:5532016.
View Article : Google Scholar : PubMed/NCBI
|