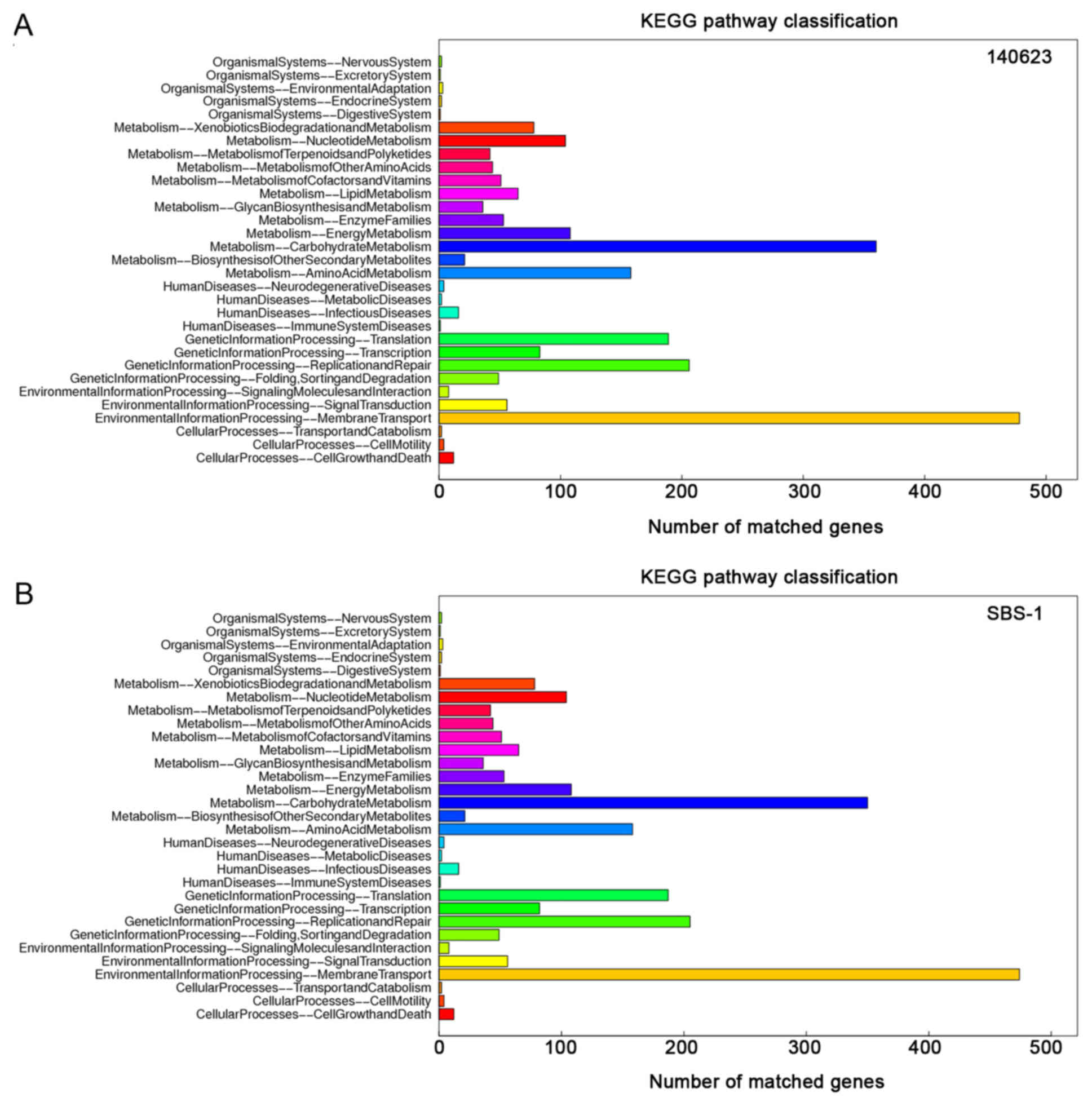
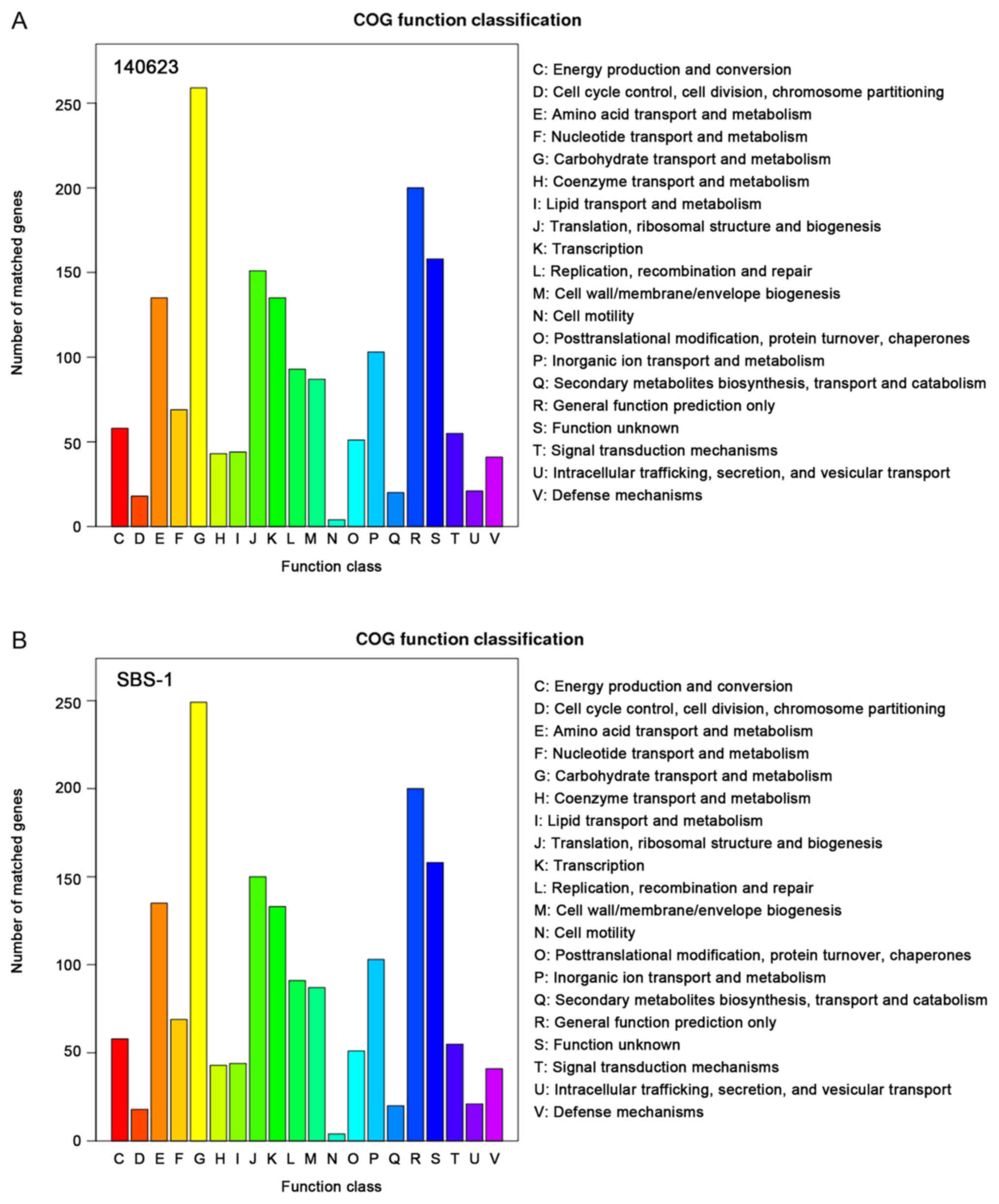
|
1
|
Rajilić-Stojanović M and de Vos WM: The
first 1000 cultured species of the human gastrointestinal
microbiota. FEMS Microbiol Rev. 38:996–1047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fisher K and Phillips C: The ecology,
epidemiology and virulence of Enterococcus. Microbiology.
155:1749–1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giannenas I, Papadopoulos E, Tsalie E,
Triantafillou E, Henikl S, Teichmann K and Tontis D: A
microbiological investigation on probiotic preparations used for
animal feeding. Microbi Res. 151:167–175. 1996. View Article : Google Scholar
|
|
4
|
Klein G: Taxonomy, ecology and antibiotic
resistance of Enterococci from food and the gastro-intestinal
tract. Int J Food Microbiol. 88:123–131. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schleifer KH and Kilpper-Bälz R: Transfer
of Streptococcus faecalis and Streptococcus faecium
to the Genus Enterococcus nom. rev. as Enterococcus
faecalis comb. nov. and E. faecium comb. nov. Int J Syst
Bacteriol. 34:31–34. 1984. View Article : Google Scholar
|
|
6
|
Vos P, Garrity G, Jones D, Krieg NR,
Ludwig W, Rainey FA, Schleifer KH and Whitman W: Bergey's Manual of
Systematic Bacteriology: Volume 3: The Firmicutes. 3. Springer
Science & Business Media; 2011
|
|
7
|
Salyers AA, Gupta A and Wang Y: Human
intestinal bacteria as reservoirs for antibiotic resistance genes.
Trends Microbiol. 12:412–416. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qin X, Galloway-Peña JR, Sillanpaa J, Roh
JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny
DM, Buhay CJ, et al: Complete genome sequence of Enterococcus
faecium strain TX16 and comparative genomic analysis of
Enterococcus faecium genomes. BMC Microbiol. 12:1352012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shanks OC, Santo Domingo JW and Graham JE:
Use of competitive DNA hybridization to identify differences in the
genomes of bacteria. J Microbiol Methods. 66:321–330. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aiba Y, Ishikawa H, Shimizu K, Noda S,
Kitada Y, Sasaki M and Koga Y: Role of internalization in the
pathogenicity of Shiga Toxin-producing Escherichia coli infection
in a gnotobiotic murine model. Microbiol Immunol. 46:723–731. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan
J, He G, Chen Y, Pan Q, Liu Y, et al: SOAPdenovo2: An empirically
improved memory-efficient short-read de novo assembler.
Gigascience. 1:182012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Altschul SF, Madden TL, Schäffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:D277–D280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M: A database for post-genome
analysis. Trends Genet. 13:375–376. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Goto S, Hattori M,
Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M and
Hirakawa M: From genomics to chemical genomics: New developments in
KEGG. Nucleic Acids Res. ((Database issue))34:D354–D357. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Magrane M and Consortium U: UniProt
Knowledgebase: A hub of integrated protein data. Database (Oxford).
2011:bar0092011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tatusov RL, Koonin EV and Lipman DJ: A
genomic perspective on protein families. Science. 278:631–637.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tatusov RL, Fedorova ND, Jackson JD,
Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov
SL, Nikolskaya AN, et al: The COG database: An updated version
includes eukaryotes. BMC Bioinformatics. 4:412003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kurtz S, Phillippy A, Delcher AL, Smoot M,
Shumway M, Antonescu C and Salzberg SL: Versatile and open software
for comparing large genomes. Genome Biol. 5:R122004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jehl MA, Arnold R and Rattei T:
Effective-a database of predicted secreted bacterial proteins.
Nucleic Acids Res (Database issue). 39:D591–D595. 2011. View Article : Google Scholar
|
|
22
|
Chen L, Xiong Z, Sun L, Yang J and Jin Q:
VFDB 2012 update: Toward the genetic diversity and molecular
evolution of bacterial virulence factors. Nucleic Acids Res.
40:D641–D645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu B and Pop M: ARDB-Antibiotic
resistance genes database. Nucleic Acids Res. 37:D443–D447. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benson G: Tandem repeats finder: A program
to analyze DNA sequences. Nucleic Acids Res. 27:573–580. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gardner PP, Daub J, Tate JG, Nawrocki EP,
Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S,
Eddy SR and Bateman A: Rfam: Updates to the RNA families database.
Nucleic Acids Res. 37:D136–D140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lagesen K, Hallin P, Rødland EA,
Staerfeldt HH, Rognes T and Ussery DW: RNAmmer: Consistent and
rapid annotation of ribosomal RNA genes. Nucleic Acids Res.
35:3100–3108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lowe TM and Eddy SR: tRNAscan-SE: A
program for improved detection of transfer RNA genes in genomic
sequence. Nucleic Acids Res. 25:955–964. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Manero A and Blanch AR: Identification of
Enterococcus spp. with a biochemical key. Appl Environ
Microbiol. 65:4425–4430. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sakayori Y, Muramatsu M, Hanada S,
Kamagata Y, Kawamoto S and Shima J: Characterization of E.
faecium mutants resistant to mundticin KS, a class IIa
bacteriocin. Microbiology. 149:2901–2908. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong Z, Zhang W, Song Y, Liu W, Xu H, Xi
X, Menghe B, Zhang H and Sun Z: Comparative genomic analysis of the
genus Enterococcus. Microbiol Res. 196:95–105. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lam MM, Seemann T, Bulach DM, Gladman SL,
Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, et
al: Comparative analysis of the first complete E. faecium
genome. J Bacteriol. 194:2334–2341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Beukers AG, Zaheer R, Goji N, Amoako KK,
Chaves AV, Ward MP and McAllister TA: Comparative genomics of
Enterococcus spp. isolated from bovine feces. BMC Microbiol.
17:522017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie QN, Jia LJ, Wang YZ, Song RT and Tang
WH: High-resolution gene profiling of infection process indicates
serine metabolism adaptation of Fusarium graminearum in
host. Sci Bull. 62:758–760. 2017. View Article : Google Scholar
|
|
34
|
Waters B and Davies J: Amino acid
variation in the GyrA subunit of bacteria potentially associated
with natural resistance to fluoroquinolone antibiotics. Antimicrob
Agents Chemother. 41:2766–2769. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dimitrijevic L, Puppo A and Rigaud J:
Superoxide dismutase activities in Rhizobium phaseoli
bacteria and bacteroids. Arch Microbiol. 139:174–178. 1984.
View Article : Google Scholar
|